Filter
Associated Lab
- Ahrens Lab (7) Apply Ahrens Lab filter
- Freeman Lab (2) Apply Freeman Lab filter
- Harris Lab (2) Apply Harris Lab filter
- Jayaraman Lab (2) Apply Jayaraman Lab filter
- Keller Lab (1) Apply Keller Lab filter
- Remove Looger Lab filter Looger Lab
- Podgorski Lab (1) Apply Podgorski Lab filter
- Schreiter Lab (3) Apply Schreiter Lab filter
- Svoboda Lab (3) Apply Svoboda Lab filter
- Turner Lab (1) Apply Turner Lab filter
- Zlatic Lab (1) Apply Zlatic Lab filter
Associated Project Team
Publication Date
Type of Publication
7 Publications
Showing 1-7 of 7 resultsCalcium imaging with protein-based indicators is widely used to follow neural activity in intact nervous systems, but current protein sensors report neural activity at timescales much slower than electrical signalling and are limited by trade-offs between sensitivity and kinetics. Here we used large-scale screening and structure-guided mutagenesis to develop and optimize several fast and sensitive GCaMP-type indicators. The resulting 'jGCaMP8' sensors, based on the calcium-binding protein calmodulin and a fragment of endothelial nitric oxide synthase, have ultra-fast kinetics (half-rise times of 2 ms) and the highest sensitivity for neural activity reported for a protein-based calcium sensor. jGCaMP8 sensors will allow tracking of large populations of neurons on timescales relevant to neural computation.
Current techniques for monitoring GABA (γ-aminobutyric acid), the primary inhibitory neurotransmitter in vertebrates, cannot follow transients in intact neural circuits. To develop a GABA sensor, we applied the design principles used to create the fluorescent glutamate receptor iGluSnFR. We used a protein derived from a previously unsequenced Pseudomonas fluorescens strain and performed structure-guided mutagenesis and library screening to obtain intensity-based GABA sensing fluorescence reporter (iGABASnFR) variants. iGABASnFR is genetically encoded, detects GABA release evoked by electric stimulation of afferent fibers in acute brain slices and produces readily detectable fluorescence increases in vivo in mice and zebrafish. We applied iGABASnFR to track mitochondrial GABA content and its modulation by an anticonvulsant, swimming-evoked, GABA-mediated transmission in zebrafish cerebellum, GABA release events during interictal spikes and seizures in awake mice, and found that GABA-mediated tone decreases during isoflurane anesthesia.
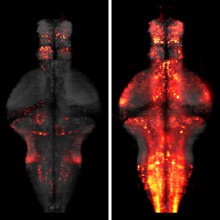
When a behavior repeatedly fails to achieve its goal, animals often give up and become passive, which can be strategic for preserving energy or regrouping between attempts. It is unknown how the brain identifies behavioral failures and mediates this behavioral-state switch. In larval zebrafish swimming in virtual reality, visual feedback can be withheld so that swim attempts fail to trigger expected visual flow. After tens of seconds of such motor futility, animals became passive for similar durations. Whole-brain calcium imaging revealed noradrenergic neurons that responded specifically to failed swim attempts and radial astrocytes whose calcium levels accumulated with increasing numbers of failed attempts. Using cell ablation and optogenetic or chemogenetic activation, we found that noradrenergic neurons progressively activated brainstem radial astrocytes, which then suppressed swimming. Thus, radial astrocytes perform a computation critical for behavior: they accumulate evidence that current actions are ineffective and consequently drive changes in behavioral states.
Genetically encoded calcium indicators (GECIs) allow measurement of activity in large populations of neurons and in small neuronal compartments, over times of milliseconds to months. Although GFP-based GECIs are widely used for in vivo neurophysiology, GECIs with red-shifted excitation and emission spectra have advantages for in vivo imaging because of reduced scattering and absorption in tissue, and a consequent reduction in phototoxicity. However, current red GECIs are inferior to the state-of-the-art GFP-based GCaMP6 indicators for detecting and quantifying neural activity. Here we present improved red GECIs based on mRuby (jRCaMP1a, b) and mApple (jRGECO1a), with sensitivity comparable to GCaMP6. We characterized the performance of the new red GECIs in cultured neurons and in mouse, Drosophila, zebrafish and C. elegans in vivo. Red GECIs facilitate deep-tissue imaging, dual-color imaging together with GFP-based reporters, and the use of optogenetics in combination with calcium imaging.
The identification of active neurons and circuits in vivo is a fundamental challenge in understanding the neural basis of behavior. Genetically encoded calcium (Ca(2+)) indicators (GECIs) enable quantitative monitoring of cellular-resolution activity during behavior. However, such indicators require online monitoring within a limited field of view. Alternatively, post hoc staining of immediate early genes (IEGs) indicates highly active cells within the entire brain, albeit with poor temporal resolution. We designed a fluorescent sensor, CaMPARI, that combines the genetic targetability and quantitative link to neural activity of GECIs with the permanent, large-scale labeling of IEGs, allowing a temporally precise "activity snapshot" of a large tissue volume. CaMPARI undergoes efficient and irreversible green-to-red conversion only when elevated intracellular Ca(2+) and experimenter-controlled illumination coincide. We demonstrate the utility of CaMPARI in freely moving larvae of zebrafish and flies, and in head-fixed mice and adult flies.
The processing of sensory input and the generation of behavior involves large networks of neurons, which necessitates new technology for recording from many neurons in behaving animals. In the larval zebrafish, light-sheet microscopy can be used to record the activity of almost all neurons in the brain simultaneously at single-cell resolution. Existing implementations, however, cannot be combined with visually driven behavior because the light sheet scans over the eye, interfering with presentation of controlled visual stimuli. Here we describe a system that overcomes the confounding eye stimulation through the use of two light sheets and combines whole-brain light-sheet imaging with virtual reality for fictively behaving larval zebrafish.
Understanding brain function requires monitoring and interpreting the activity of large networks of neurons during behavior. Advances in recording technology are greatly increasing the size and complexity of neural data. Analyzing such data will pose a fundamental bottleneck for neuroscience. We present a library of analytical tools called Thunder built on the open-source Apache Spark platform for large-scale distributed computing. The library implements a variety of univariate and multivariate analyses with a modular, extendable structure well-suited to interactive exploration and analysis development. We demonstrate how these analyses find structure in large-scale neural data, including whole-brain light-sheet imaging data from fictively behaving larval zebrafish, and two-photon imaging data from behaving mouse. The analyses relate neuronal responses to sensory input and behavior, run in minutes or less and can be used on a private cluster or in the cloud. Our open-source framework thus holds promise for turning brain activity mapping efforts into biological insights.