Filter
Associated Lab
- Aso Lab (29) Apply Aso Lab filter
- Betzig Lab (1) Apply Betzig Lab filter
- Bock Lab (2) Apply Bock Lab filter
- Branson Lab (7) Apply Branson Lab filter
- Card Lab (5) Apply Card Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Dickson Lab (2) Apply Dickson Lab filter
- Druckmann Lab (1) Apply Druckmann Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Funke Lab (1) Apply Funke Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (2) Apply Heberlein Lab filter
- Hermundstad Lab (2) Apply Hermundstad Lab filter
- Hess Lab (5) Apply Hess Lab filter
- Jayaraman Lab (5) Apply Jayaraman Lab filter
- Lippincott-Schwartz Lab (1) Apply Lippincott-Schwartz Lab filter
- Looger Lab (2) Apply Looger Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Reiser Lab (14) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Romani Lab (1) Apply Romani Lab filter
- Remove Rubin Lab filter Rubin Lab
- Saalfeld Lab (4) Apply Saalfeld Lab filter
- Scheffer Lab (7) Apply Scheffer Lab filter
- Schreiter Lab (1) Apply Schreiter Lab filter
- Simpson Lab (3) Apply Simpson Lab filter
- Singer Lab (1) Apply Singer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Stern Lab (1) Apply Stern Lab filter
- Svoboda Lab (3) Apply Svoboda Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Truman Lab (4) Apply Truman Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
- Turner Lab (5) Apply Turner Lab filter
- Zuker Lab (1) Apply Zuker Lab filter
Associated Project Team
Publication Date
- 2024 (2) Apply 2024 filter
- 2023 (6) Apply 2023 filter
- 2022 (1) Apply 2022 filter
- 2021 (4) Apply 2021 filter
- 2020 (9) Apply 2020 filter
- 2019 (6) Apply 2019 filter
- 2018 (7) Apply 2018 filter
- 2017 (15) Apply 2017 filter
- 2016 (3) Apply 2016 filter
- 2015 (16) Apply 2015 filter
- 2014 (9) Apply 2014 filter
- 2013 (5) Apply 2013 filter
- 2012 (8) Apply 2012 filter
- 2011 (4) Apply 2011 filter
- 2010 (4) Apply 2010 filter
- 2009 (2) Apply 2009 filter
- 2008 (4) Apply 2008 filter
- 2007 (2) Apply 2007 filter
- 2006 (1) Apply 2006 filter
- 2002 (1) Apply 2002 filter
- 2000 (2) Apply 2000 filter
- 1999 (1) Apply 1999 filter
- 1997 (1) Apply 1997 filter
- 1995 (2) Apply 1995 filter
- 1994 (2) Apply 1994 filter
- 1993 (2) Apply 1993 filter
- 1992 (1) Apply 1992 filter
- 1991 (2) Apply 1991 filter
- 1990 (3) Apply 1990 filter
- 1989 (2) Apply 1989 filter
- 1987 (2) Apply 1987 filter
- 1986 (1) Apply 1986 filter
- 1985 (1) Apply 1985 filter
- 1984 (1) Apply 1984 filter
- 1983 (1) Apply 1983 filter
- 1982 (2) Apply 1982 filter
- 1981 (1) Apply 1981 filter
- 1979 (1) Apply 1979 filter
- 1973 (1) Apply 1973 filter
Type of Publication
138 Publications
Showing 131-138 of 138 resultsUsing a novel method for detecting cross-homologous nucleic acid sequences we have isolated the gene coding for the major rhodopsin of Drosophila melanogaster and mapped it to chromosomal region 92B8-11. Comparison of cDNA and genomic DNA sequences indicates that the gene is divided into five exons. The amino acid sequence deduced from the nucleotide sequence is 373 residues long, and the polypeptide chain contains seven hydrophobic segments that appear to correspond to the seven transmembrane segments characteristic of other rhodopsins. Three regions of Drosophila rhodopsin are highly conserved with the corresponding domains of bovine rhodopsin, suggesting an important role for these polypeptide regions.
We have made a P-element derivative called Pc[ry], which carries the selectable marker gene rosy, but which acts like a nondefective, intact P element. It transposes autonomously into the germline chromosomes of an M-strain Drosophila embryo and it mobilizes in trans the defective P elements of the singed-weak allele. Frameshift mutations introduced into any of the four major open reading frames of the P sequence were each sufficient to eliminate the transposase activity, but none affected signals required in cis for transposition of the element. Complementation tests between pairs of mutant elements suggest that a single polypeptide comprises the transposase. We have examined transcripts of P elements both from natural P strains and from lines containing only nondefective Pc[ry] elements, and have identified two RNA species that appear to be specific for autonomous elements.
Thirty-six isogenic D. melanogaster strains that differed only in the chromosomal location of a 7.2 or an 8.1 kb DNA segment containing the (autosomal) rosy gene were constructed by P-element-mediated gene transfer. Since the flies were homozygous for a rosy- allele, rosy gene function in these indicated the influence of flanking sequences on gene expression. The tissue distribution of XDH activity in all the strains was normal. Each line exhibited a characteristic level of adult XDH-specific activity. The majority of these values were close to wild-type levels; however, the total variation in specific activity among the lines was nearly fivefold. Thus position effects influence expression of the rosy gene quantitatively but do not detectably alter tissue specificity. X-linked rosy insertions were expressed on average 1.6 times more activity in males than in females. Hence the gene acquires at least partial dosage compensation upon insertion into the X chromosome.
Exogenous DNA sequences were introduced into the Drosophila germ line. A rosy transposon (ry1), constructed by inserting a chromosomal DNA fragment containing the wild-type rosy gene into a P transposable element, transformed germ line cells in 20 to 50 percent of the injected rosy mutant embryos. Transformants contained one or two copies of chromosomally integrated, intact ry1 that were stably inherited in subsequent generations. These transformed flies had wild-type eye color indicating that the visible genetic defect in the host strain could be fully and permanently corrected by the transferred gene. To demonstrate the generality of this approach, a DNA segment that does not confer a recognizable phenotype on recipients was also transferred into germ line chromosomes.
We have shown previously that four of five white mutant alleles arising in P-M dysgenic hybrids result from the insertion of strongly homologous DNA sequence elements. We have named these P elements. We report that P elements are present in 30-50 copies per haploid genome in all P strains examined and apparently are missing entirely from all M strains examined, with one exception. Furthermore, members of the P family apparently transpose frequently in P-M dysgenic hybrids; chromosomes descendant from P-M dysgenic hybrids frequently show newly acquired P elements. Finally, the strain-specific breakpoint hotspots for the rearrangement of the pi 2 P X chromosome occurring in P-M dysgenic hybrids are apparently sites of residence of P elements. These observations strongly support the P factor hypothesis for the mechanistic basis of P-M hybrid dysgenesis.
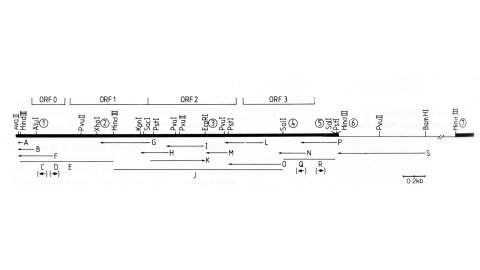
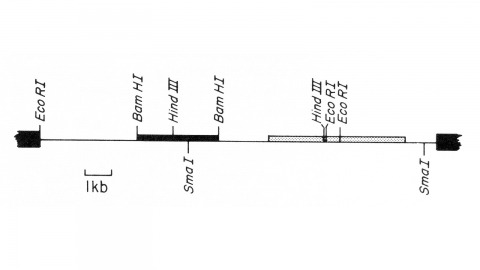
We describe the isolation of a cloned DNA segment carrying unique sequences from the white locus of Drosophila melanogaster. Sequences within the cloned segment are shown to hybridize in situ to the white locus region on the polytene chromosomes of both wild-type strains and strains carrying chromosomal rearrangements whose breakpoints bracket the white locus. We further show that two small deficiency mutations, deleting white locus genetic elements but not those of complementation groups contiguous to white, delete the genomic sequences corresponding to a portion of the cloned segment. The strategy we have employed to isolate this cloned segment exploits the existence of an allele at the white locus containing a copy of a previously cloned transposable, reiterated DNA sequence element. We describe a simple, rapid method for retrieving cloned segments carrying a copy of the transposable element together with contiguous sequences corresponding to this allele. The strategy described is potentially general and we discuss its application to the cloning of the DNA sequences of other genes in Drosophila, including those identified only by genetic analysis and for which no RNA product is known.
The stability of elements of three different dispersed repeated gene families in the genome of Drosophila tissue culture cells has been examined. Different amounts of sequences homologous to elements of 412, copia and 297 dispersed repeated gene families are found in the genomes of D. melanogaster embryonic and tissue culture cells. In general the amount of these sequences is increased in the cell lines. The additional sequences homologous to 412, copia and 297 occur as intact elements and are dispersed to new sites in the cell culture genome. It appears that these elements can insert at many alternative sites. We also describe a DNA sequence arrangement found in the D. melanogaster embryo genome which appears to result from a transposition of an element of the copia dispersed repeated gene family into a new chromosomal site. The mechanism of insertion of this copia element is precise to within 90 bp and may involve a region of weak sequence homology between the site of insertion and the direct terminal repeats of the copia element.