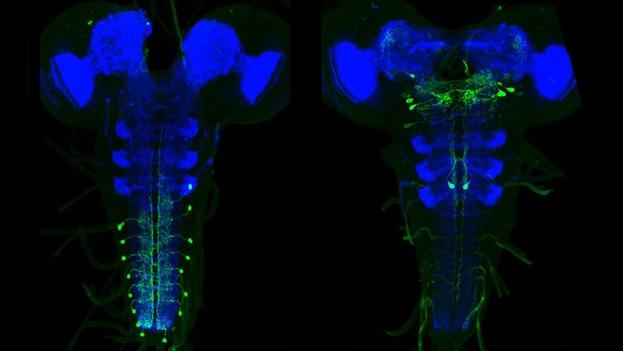
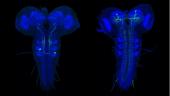
The larval nervous system contains 10,000 neurons, about 2,000 of which are in the brain with the remainder in the ventral nervous system. Although there are far fewer cells in the larval nervous system than in the adult CNS, most neural cell types are nevertheless present in the larva, with the notable exceptions of neurons of the optic lobes and the central complex. In the adult, there may be many neurons in a given cell class whereas the same class in the larva may consist of a single cell. Consequently, the larval CNS presents a brain of high complexity but with numerical simplicity. Catalyzed by Albert Cardona’s group at Janelia, the complete connectome of the first instar larva is being reconstructed from EM sections (using a primary dataset generated by Rick Fetter and the FlyEM project). Through a growing, web-based international collaboration that now involves twelve laboratories, the reconstruction is 26% finished and on track to be completed within two years. It will be only the second example of a fully reconstructed CNS, and contains about 30 times more neurons than that from Caenorhabditis elegans. The larval work in FlyLight is coordinated with this EM effort. Our light-level information on the shapes of neurons aids the EM reconstruction effort. More importantly, this information provides a method to move from an EM reconstructed neuron to a genetic line that allows that neuron to be studied physiologically and behaviorally. The larval work of FlyLight also aids the behavioral screens of the Larval Olympiad Project, a consortium of 10 labs doing high throughput screens on behaviors including feeding, escape locomotion, navigation, and classical conditioning.
The larval expression patterns of 7,185 of the Generation 1 GAL4 lines at Janelia were initially characterized and representative images and stacks from each line have been publicly available since May 2013. This website includes a database that allows the community to search for lines that have notable expression patterns in particular regions of the CNS or that show expression in particular cell types.
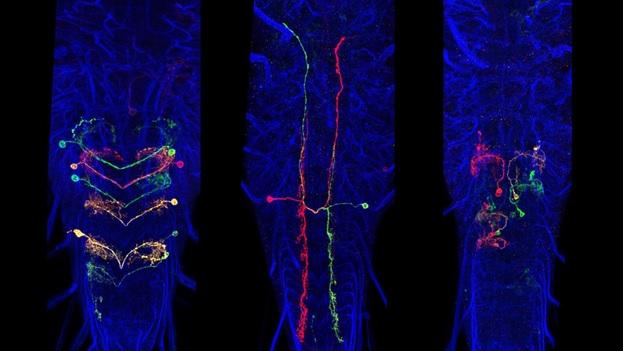
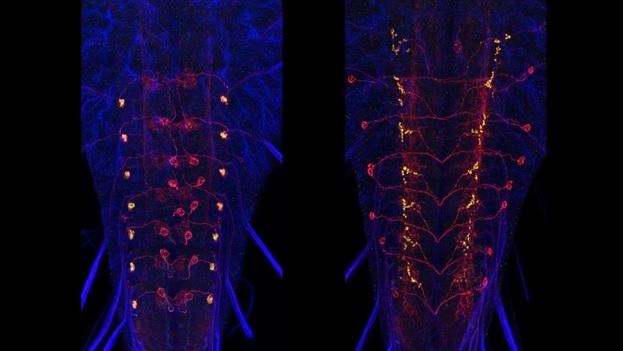
A subset of Generation 1 lines was selected based on sparse expression in particular regions of the CNS and these provided the material for the behavioral screens of the Larval Olympiad Project. About 1,400 of these sparse lines were subjected to multicolor flip-out analysis (MCFO) to characterize the neurons present in each. For this analysis, the CNS was divided into four regions: the brain and the subesophageal, thoracic, and abdominal neuromeres. Confocal stacks were taken at 63x at 0.5-micron intervals, and single neurons were reconstructed in dorsal, transverse and lateral views and related to landmarks in the neuropil. We have flipped out and imaged over 10,000 nervous systems from 1,500 sparse lines. To date, over 20,000 neurons have been scored and placed in 1,353 neuronal classes. The number of neuron classes for the various regions is currently: brain: 615; subesophageal ganglia: 147; thoracic ganglia: 205; abdominal ganglia: 386 neurons. Although we have abundant flip-out material for all of the CNS regions, our work on identifying and describing new neurons has concentrated on the brain and abdominal neuromeres because these regions are the current focus of the EM reconstructions and the Larval Olympiad studies. We estimate that we have identified about 60-70% of the neuron types in the brain and over 95% of the neuron types in the abdominal neuromeres.
Data from the neurons in each sparse line are entered in an “Identified Neuron” database that is used to search either for the neurons that express in a specific line or for the lines whose expression includes a specific neuron. These data were used to determine which Generation 1 lines might be useful as a split-GAL4 component [either AD, DBD, or both], and to predict which spilt GAL4 intersections would most likely target a particular neuron. Additional intersection options came with Barry Dickson’s arrival at Janelia in October 2013. He brought a collection of 2,800 new GAL4 lines that were also configured into split-GAL4 halves. Their expression in the adult central brain was known, but not their larval patterns. We have completed the imaging of the larval CNSs for 2,871 of these lines at 40X magnification. We subsequently did MCFO analysis on 326 of the most promising lines to facilitate designing split combinations.
Examples of intersection combinations that target input and output neurons to the various zones of the medial lobe of the larval mushroom body.
Completion of Larval Neuron Annotation and Establishment of a Larval Neuron Database
Over the next year or two, we will be finishing the annotation of the neurons in the subesophageal and thoracic regions of the larval CNS. The data from all of the neurons will be combined in a searchable website. As the EM project comes to completion, there will be links from each neuron to its EM profile and also links to the various GAL4 and LexA lines that contain the cell.
Coordination of Split-GAL4 Line Production with the Growing EM Connectome
The EM reconstruction of the larval CNS is over 30% finished with some areas, such as the mushroom bodies and the optic and olfactory lobes, being essentially complete. Our selection of target neurons for the split-GAL4 lines was largely focused on CNS areas and pathways that were being reconstructed in the EM. As the EM project moves away from both sensory and motor regions, researchers are identifying third and fourth order interneurons that are highly connected to interneurons of interest. We intend to target these neurons for production of additional stable split lines so that the physiological properties and their behavioral roles of these higher order cells can also be studied.
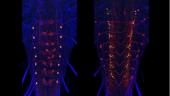
After we find split-GAL4 intersections that give us a desired neuron, a homozygous stock that contains both transgenes is constructed. These stable lines can then be used to study the specific cells of interest. The expression of the stable stocks needs to be confirmed. The flies will be crossed to both a membrane marker and a synaptic vesicle marker (Syb::GFP) to provide good overall anatomy and synaptic output sites. Imaging will be done at 40x and projections and stacks placed in an open access website. Unlike in the adult, we generally do not require MCFO to analyze the stable lines due to the smaller number of cells in the larval nervous system.
Polarity Markers for the Local Interneurons
For both the brain and the ventral neuromeres, projection neurons that run through the tracts and commissures show a distinctive polarity with bulbous presynaptic terminals and fine, filamentous postsynaptic processes. These designations of pre- and postsynaptic portions of the cell are repeatedly confirmed when compared to the ground-truth of the cell’s EM reconstruction. Input and output sites are much harder to determine, though, for the mixed arbors of the local interneurons. We are using membrane and polarity markers, as described above for the adult, to characterize the input and output regions of these local interneurons in the larva. These experiments use selected split-GAL4 lines and the most appropriate Generation 1 lines.