Filter
Associated Lab
Publication Date
- Remove 2010-12-31 19:00 – 2011-12-31 19:00 filter 2010-12-31 19:00 – 2011-12-31 19:00
- September 16, 2011 (1) Apply September 16, 2011 filter
- September 8, 2011 (1) Apply September 8, 2011 filter
- September 2, 2011 (1) Apply September 2, 2011 filter
- September 1, 2011 (5) Apply September 1, 2011 filter
- Remove September 2011 filter September 2011
8 Janelia Publications
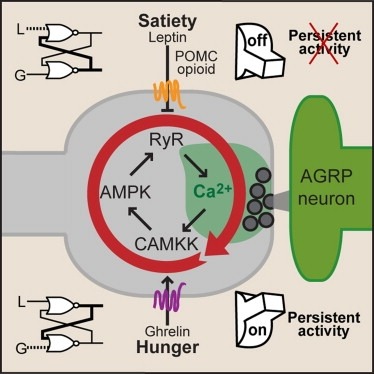
Showing 1-8 of 8 resultsSynaptic plasticity in response to changes in physiologic state is coordinated by hormonal signals across multiple neuronal cell types. Here, we combine cell-type-specific electrophysiological, pharmacological, and optogenetic techniques to dissect neural circuits and molecular pathways controlling synaptic plasticity onto AGRP neurons, a population that regulates feeding. We find that food deprivation elevates excitatory synaptic input, which is mediated by a presynaptic positive feedback loop involving AMP-activated protein kinase. Potentiation of glutamate release was triggered by the orexigenic hormone ghrelin and exhibited hysteresis, persisting for hours after ghrelin removal. Persistent activity was reversed by the anorexigenic hormone leptin, and optogenetic photostimulation demonstrated involvement of opioid release from POMC neurons. Based on these experiments, we propose a memory storage device for physiological state constructed from bistable synapses that are flipped between two sustained activity states by transient exposure to hormones signaling energy levels.
A specialist neuron uses an intriguing process to help control the body's response to hunger. A lipid pathway involving the breakdown of cellular components regulates the expression of a neuropeptide that affects feeding and body weight.
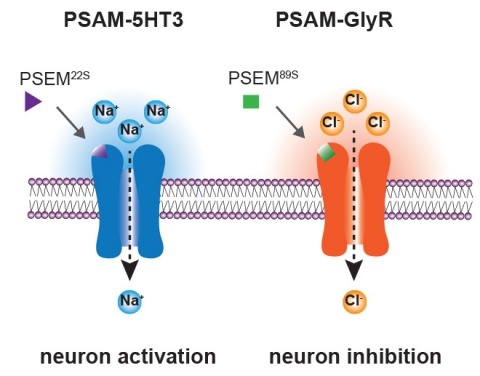
Ionic flux mediates essential physiological and behavioral functions in defined cell populations. Cell type-specific activators of diverse ionic conductances are needed for probing these effects. We combined chemistry and protein engineering to enable the systematic creation of a toolbox of ligand-gated ion channels (LGICs) with orthogonal pharmacologic selectivity and divergent functional properties. The LGICs and their small-molecule effectors were able to activate a range of ionic conductances in genetically specified cell types. LGICs constructed for neuronal perturbation could be used to selectively manipulate neuron activity in mammalian brains in vivo. The diversity of ion channel tools accessible from this approach will be useful for examining the relationship between neuronal activity and animal behavior, as well as for cell biological and physiological applications requiring chemical control of ion conductance.
Concomitant with the publication of this Special Issue of Neuroinformatics, a substantially updated version of the DIADEM web site has been released at http://diademchallenge.org. This web site was originally designed to host the challenge for automating the digital reconstruction of axonal and dendritic morphology (hence the DIADEM acronym). This post-competition version features additional content for continued use as the access point for DIADEM-related material. From the very beginning, one of the spirits of DIADEM has been to share data and resources with the neuroscience research community at large. The resources available from or linked to the DIADEM website constitute a substantial scientific legacy of the 2009/2010 competition. The new content includes finalist algorithms, image stack data, gold standard reconstructions, an updated DIADEM metric, and a retrospective on the competition in text and images.
We took advantage of the unusual genomic organization of the ciliate Oxytricha trifallax to screen for eukaryotic non-coding RNA (ncRNA) genes. Ciliates have two types of nuclei: a germ line micronucleus that is usually transcriptionally inactive, and a somatic macronucleus that contains a reduced, fragmented and rearranged genome that expresses all genes required for growth and asexual reproduction. In some ciliates including Oxytricha, the macronuclear genome is particularly extreme, consisting of thousands of tiny ’nanochromosomes’, each of which usually contains only a single gene. Because the organism itself identifies and isolates most of its genes on single-gene nanochromosomes, nanochromosome structure could facilitate the discovery of unusual genes or gene classes, such as ncRNA genes. Using a draft Oxytricha genome assembly and a custom-written protein-coding genefinding program, we identified a subset of nanochromosomes that lack any detectable protein-coding gene, thereby strongly enriching for nanochromosomes that carry ncRNA genes. We found only a small proportion of non-coding nanochromosomes, suggesting that Oxytricha has few independent ncRNA genes besides homologs of already known RNAs. Other than new members of known ncRNA classes including C/D and H/ACA snoRNAs, our screen identified one new family of small RNA genes, named the Arisong RNAs, which share some of the features of small nuclear RNAs.
Because of its genetic, molecular, and behavioral tractability, Drosophila has emerged as a powerful model system for studying molecular and cellular mechanisms underlying the development and function of nervous systems. The Drosophila nervous system has fewer neurons and exhibits a lower glia:neuron ratio than is seen in vertebrate nervous systems. Despite the simplicity of the Drosophila nervous system, glial organization in flies is as sophisticated as it is in vertebrates. Furthermore, fly glial cells play vital roles in neural development and behavior. In addition, powerful genetic tools are continuously being created to explore cell function in vivo. In taking advantage of these features, the fly nervous system serves as an excellent model system to study general aspects of glial cell development and function in vivo. In this article, we review and discuss advanced genetic tools that are potentially useful for understanding glial cell biology in Drosophila.