Filter
Associated Lab
Associated Project Team
- Remove FlyLight filter FlyLight
Publication Date
3 Janelia Publications
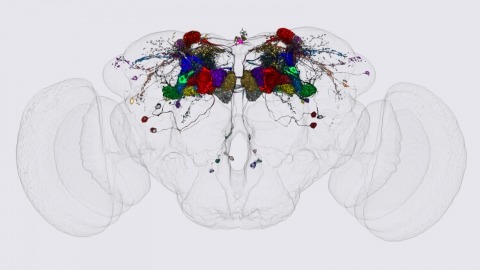
Showing 1-3 of 3 resultsWe identified the neurons comprising the Drosophila mushroom body (MB), an associative center in invertebrate brains, and provide a comprehensive map describing their potential connections. Each of the 21 MB output neuron (MBON) types elaborates segregated dendritic arbors along the parallel axons of ∼2000 Kenyon cells, forming 15 compartments that collectively tile the MB lobes. MBON axons project to five discrete neuropils outside of the MB and three MBON types form a feedforward network in the lobes. Each of the 20 dopaminergic neuron (DAN) types projects axons to one, or at most two, of the MBON compartments. Convergence of DAN axons on compartmentalized Kenyon cell-MBON synapses creates a highly ordered unit that can support learning to impose valence on sensory representations. The elucidation of the complement of neurons of the MB provides a comprehensive anatomical substrate from which one can infer a functional logic of associative olfactory learning and memory.
We report the larval CNS expression patterns for 6,650 GAL4 lines based on cis-regulatory regions (CRMs) from the Drosophila genome. Adult CNS expression patterns were previously reported for this collection, thereby providing a unique resource for determining the origins of adult cells. An illustrative example reveals the origin of the astrocyte-like glia of the ventral CNS. Besides larval neurons and glia, the larval CNS contains scattered lineages of immature, adult-specific neurons. Comparison of lineage expression within this large collection of CRMs provides insight into the codes used for designating neuronal types. The CRMs encode both dense and sparse patterns of lineage expression. There is little correlation between brain and thoracic lineages in patterns of sparse expression, but expression in the two regions is highly correlated in the dense mode. The optic lobes, by comparison, appear to use a different set of genetic instructions in their development.
The Drosophila cerebrum originates from about 100 neuroblasts per hemisphere, with each neuroblast producing a characteristic set of neurons. Neurons from a neuroblast are often so diverse that many neuron types remain unexplored. We developed new genetic tools that target neuroblasts and their diverse descendants, increasing our ability to study fly brain structure and development. Common enhancer-based drivers label neurons on the basis of terminal identities rather than origins, which provides limited labeling in the heterogeneous neuronal lineages. We successfully converted conventional drivers that are temporarily expressed in neuroblasts, into drivers expressed in all subsequent neuroblast progeny. One technique involves immortalizing GAL4 expression in neuroblasts and their descendants. Another depends on loss of the GAL4 repressor, GAL80, from neuroblasts during early neurogenesis. Furthermore, we expanded the diversity of MARCM-based reagents and established another site-specific mitotic recombination system. Our transgenic tools can be combined to map individual neurons in specific lineages of various genotypes.