Filter
Associated Lab
- Card Lab (1) Apply Card Lab filter
- Cui Lab (1) Apply Cui Lab filter
- Gonen Lab (1) Apply Gonen Lab filter
- Hess Lab (1) Apply Hess Lab filter
- Lavis Lab (1) Apply Lavis Lab filter
- Looger Lab (2) Apply Looger Lab filter
- Rubin Lab (1) Apply Rubin Lab filter
- Svoboda Lab (1) Apply Svoboda Lab filter
- Truman Lab (1) Apply Truman Lab filter
Publication Date
- April 26, 2012 (1) Apply April 26, 2012 filter
- April 17, 2012 (1) Apply April 17, 2012 filter
- April 15, 2012 (1) Apply April 15, 2012 filter
- April 9, 2012 (1) Apply April 9, 2012 filter
- April 5, 2012 (1) Apply April 5, 2012 filter
- April 3, 2012 (1) Apply April 3, 2012 filter
- April 1, 2012 (3) Apply April 1, 2012 filter
- Remove April 2012 filter April 2012
- Remove 2012 filter 2012
9 Janelia Publications
Showing 1-9 of 9 resultsThe mechanisms linking sensation and action during learning are poorly understood. Layer 2/3 neurons in the motor cortex might participate in sensorimotor integration and learning; they receive input from sensory cortex and excite deep layer neurons, which control movement. Here we imaged activity in the same set of layer 2/3 neurons in the motor cortex over weeks, while mice learned to detect objects with their whiskers and report detection with licking. Spatially intermingled neurons represented sensory (touch) and motor behaviours (whisker movements and licking). With learning, the population-level representation of task-related licking strengthened. In trained mice, population-level representations were redundant and stable, despite dynamism of single-neuron representations. The activity of a subpopulation of neurons was consistent with touch driving licking behaviour. Our results suggest that ensembles of motor cortex neurons couple sensory input to multiple, related motor programs during learning.
Microscopic images of specific proteins in their cellular context yield important insights into biological processes and cellular architecture. The advent of superresolution optical microscopy techniques provides the possibility to augment EM with nanometer-resolution fluorescence microscopy to access the precise location of proteins in the context of cellular ultrastructure. Unfortunately, efforts to combine superresolution fluorescence and EM have been stymied by the divergent and incompatible sample preparation protocols of the two methods. Here, we describe a protocol that preserves both the delicate photoactivatable fluorescent protein labels essential for superresolution microscopy and the fine ultrastructural context of EM. This preparation enables direct 3D imaging in 500- to 750-nm sections with interferometric photoactivatable localization microscopy followed by scanning EM images generated by focused ion beam ablation. We use this process to "colorize" detailed EM images of the mitochondrion with the position of labeled proteins. The approach presented here has provided a new level of definition of the in vivo nature of organization of mitochondrial nucleoids, and we expect this straightforward method to be applicable to many other biological questions that can be answered by direct imaging.
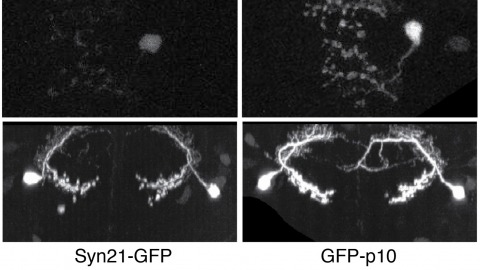
The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibire(ts1), and for the use of cytoplasmic GFP to image the fine processes of neurons.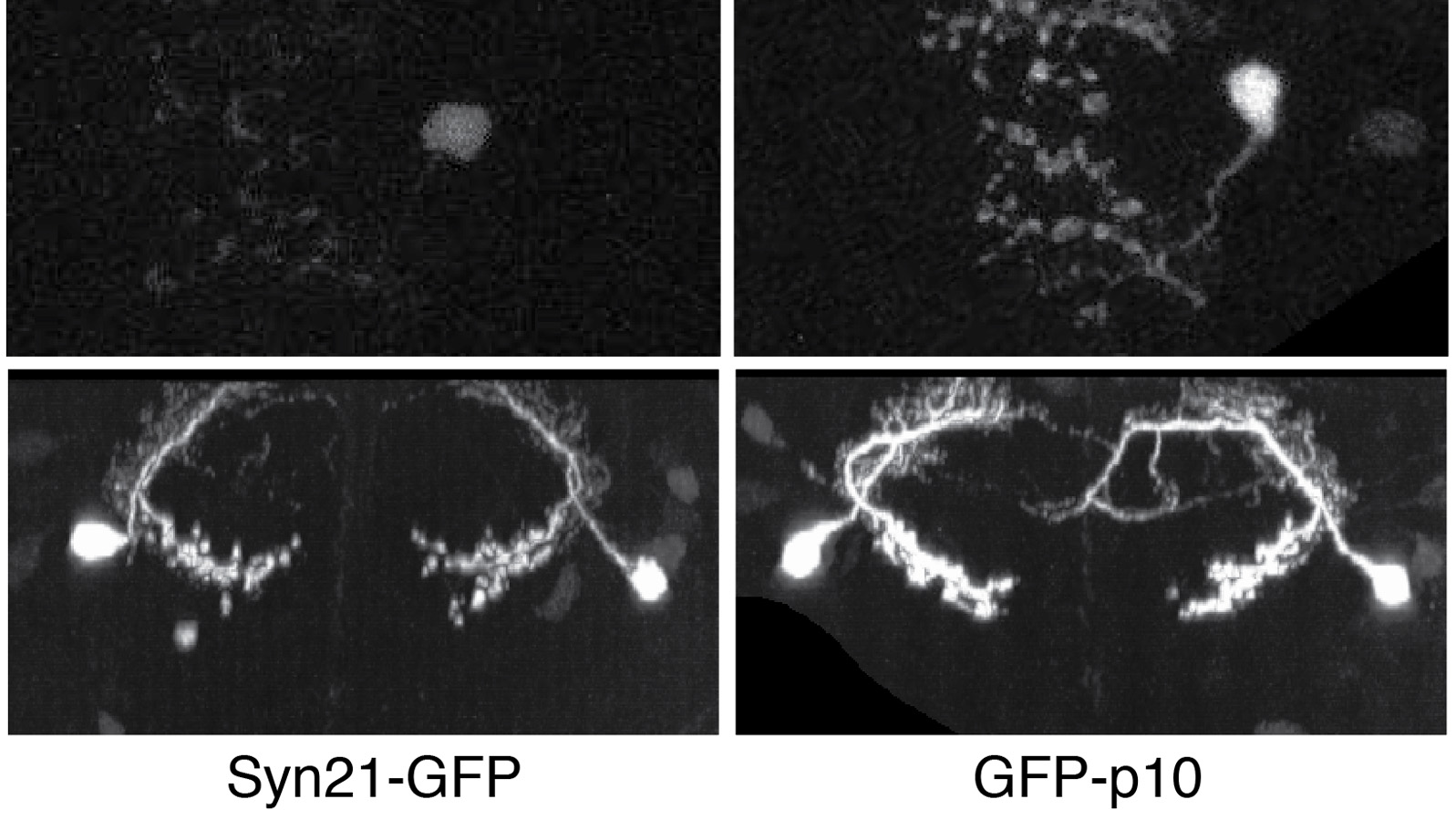
Voltage-gated ion channels are responsible for transmitting electrochemical signals in both excitable and non-excitable cells. Structural studies of voltage-gated potassium and sodium channels by X-ray crystallography have revealed atomic details on their voltage-sensor domains (VSDs) and pore domains, and were put in context of disparate mechanistic views on the voltage-driven conformational changes in these proteins. Functional investigation of voltage-gated channels in membranes, however, showcased a mechanism of lipid-dependent gating for voltage-gated channels, suggesting that the lipids play an indispensible and critical role in the proper gating of many of these channels. Structure determination of membrane-embedded voltage-gated ion channels appears to be the next frontier in fully addressing the mechanism by which the VSDs control channel opening. Currently electron crystallography is the only structural biology method in which a membrane protein of interest is crystallized within a complete lipid-bilayer mimicking the native environment of a biological membrane. At a sufficiently high resolution, an electron crystallographic structure could reveal lipids, the channel and their mutual interactions at the atomic level. Electron crystallography is therefore a promising avenue toward understanding how lipids modulate channel activation through close association with the VSDs.
Previous implementations of structured-illumination microscopy (SIM) were slow or designed for one-color excitation, sacrificing two unique and extremely beneficial aspects of light microscopy: live-cell imaging in multiple colors. This is especially unfortunate because, among the resolution-extending techniques, SIM is an attractive choice for live-cell imaging; it requires no special fluorophores or high light intensities to achieve twice diffraction-limited resolution in three dimensions. Furthermore, its wide-field nature makes it light-efficient and decouples the acquisition speed from the size of the lateral field of view, meaning that high frame rates over large volumes are possible. Here, we report a previously undescribed SIM setup that is fast enough to record 3D two-color datasets of living whole cells. Using rapidly programmable liquid crystal devices and a flexible 2D grid pattern algorithm to switch between excitation wavelengths quickly, we show volume rates as high as 4 s in one color and 8.5 s in two colors over tens of time points. To demonstrate the capabilities of our microscope, we image a variety of biological structures, including mitochondria, clathrin-coated vesicles, and the actin cytoskeleton, in either HeLa cells or cultured neurons.
Since the original identification of GFP from jellyfish and corals, the genetically encoded fluorescent proteins have become mainstream indicators for imaging. Functionally homologous candidates exist in more highly evolved bioluminescent invertebrates, including echinoderms. For example, in brittlestars, stimulus-evoked bioluminescence is transient, lasting seconds, and emanates from specialized cells (photocytes). Prior to light emission, we observe little or no green fluorescence. However, concurrent with light emission, an intense green, calcium-dependent fluorescence develops that persists indefinitely.
Calcium (Ca(2+)) is a ubiquitous signaling molecule that accumulates in the cytoplasm in response to diverse classes of stimuli and, in turn, regulates many aspects of cell function. In neurons, Ca(2+) influx in response to action potentials or synaptic stimulation triggers neurotransmitter release, modulates ion channels, induces synaptic plasticity, and activates transcription. In this article, we discuss the factors that regulate Ca(2+) signaling in mammalian neurons with a particular focus on Ca(2+) signaling within dendritic spines. This includes consideration of the routes of entry and exit of Ca(2+), the cellular mechanisms that establish the temporal and spatial profile of Ca(2+) signaling, and the biophysical criteria that determine which downstream signals are activated when Ca(2+) accumulates in a spine. Furthermore, we also briefly discuss the technical advances that made possible the quantitative study of Ca(2+) signaling in dendritic spines.
Escape behaviors are, by necessity, fast and robust, making them excellent systems with which to study the neural basis of behavior. This is especially true in insects, which have comparatively tractable nervous systems and members who are amenable to manipulation with genetic tools. Recent technical developments in high-speed video reveal that, despite their short duration, insect escape behaviors are more complex than previously appreciated. For example, before initiating an escape jump, a fly performs sophisticated posture and stimulus-dependent preparatory leg movements that enable it to jump away from a looming threat. This newfound flexibility raises the question of how the nervous system generates a behavior that is both rapid and flexible. Recordings from the cricket nervous system suggest that synchrony between the activity of specific interneuron pairs may provide a rapid cue for the cricket to detect the direction of an approaching predator and thus which direction it should run. Technical advances make possible wireless recording from neurons while locusts escape from a looming threat, enabling, for the first time, a direct correlation between the activity of multiple neurons and the time-course of an insect escape behavior.