Filter
Associated Lab
Associated Project Team
8 Janelia Publications
Showing 1-8 of 8 resultsAnimals discriminate stimuli, learn their predictive value and use this knowledge to modify their behavior. In Drosophila, the mushroom body (MB) plays a key role in these processes. Sensory stimuli are sparsely represented by ∼2000 Kenyon cells, which converge onto 34 output neurons (MBONs) of 21 types. We studied the role of MBONs in several associative learning tasks and in sleep regulation, revealing the extent to which information flow is segregated into distinct channels and suggesting possible roles for the multi-layered MBON network. We also show that optogenetic activation of MBONs can, depending on cell type, induce repulsion or attraction in flies. The behavioral effects of MBON perturbation are combinatorial, suggesting that the MBON ensemble collectively represents valence. We propose that local, stimulus-specific dopaminergic modulation selectively alters the balance within the MBON network for those stimuli. Our results suggest that valence encoded by the MBON ensemble biases memory-based action selection.
We identified the neurons comprising the Drosophila mushroom body (MB), an associative center in invertebrate brains, and provide a comprehensive map describing their potential connections. Each of the 21 MB output neuron (MBON) types elaborates segregated dendritic arbors along the parallel axons of ∼2000 Kenyon cells, forming 15 compartments that collectively tile the MB lobes. MBON axons project to five discrete neuropils outside of the MB and three MBON types form a feedforward network in the lobes. Each of the 20 dopaminergic neuron (DAN) types projects axons to one, or at most two, of the MBON compartments. Convergence of DAN axons on compartmentalized Kenyon cell-MBON synapses creates a highly ordered unit that can support learning to impose valence on sensory representations. The elucidation of the complement of neurons of the MB provides a comprehensive anatomical substrate from which one can infer a functional logic of associative olfactory learning and memory.
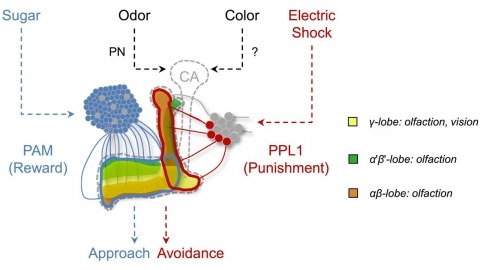
In nature, animals form memories associating reward or punishment with stimuli from different sensory modalities, such as smells and colors. It is unclear, however, how distinct sensory memories are processed in the brain. We established appetitive and aversive visual learning assays for Drosophila that are comparable to the widely used olfactory learning assays. These assays share critical features, such as reinforcing stimuli (sugar reward and electric shock punishment), and allow direct comparison of the cellular requirements for visual and olfactory memories. We found that the same subsets of dopamine neurons drive formation of both sensory memories. Furthermore, distinct yet partially overlapping subsets of mushroom body intrinsic neurons are required for visual and olfactory memories. Thus, our results suggest that distinct sensory memories are processed in a common brain center. Such centralization of related brain functions is an economical design that avoids the repetition of similar circuit motifs.
An important strategy for efficient neural coding is to match the range of cellular responses to the distribution of relevant input signals. However, the structure and relevance of sensory signals depend on behavioral state. Here, we show that behavior modifies neural activity at the earliest stages of fly vision. We describe a class of wide-field neurons that provide feedback to the most peripheral layer of the Drosophila visual system, the lamina. Using in vivo patch-clamp electrophysiology, we found that lamina wide-field neurons respond to low-frequency luminance fluctuations. Recordings in flying flies revealed that the gain and frequency tuning of wide-field neurons change during flight, and that these effects are mimicked by the neuromodulator octopamine. Genetically silencing wide-field neurons increased behavioral responses to slow-motion stimuli. Together, these findings identify a cell type that is gated by behavior to enhance neural coding by subtracting low-frequency signals from the inputs to motion detection circuits.
Visual motion perception is critical to many animal behaviors, and flies have emerged as a powerful model system for exploring this fundamental neural computation. Although numerous studies have suggested that fly motion vision is governed by a simple neural circuit [1-3], the implementation of this circuit has remained mysterious for decades. Connectomics and neurogenetics have produced a surge in recent progress, and several studies have shown selectivity for light increments (ON) or decrements (OFF) in key elements associated with this circuit [4-7]. However, related studies have reached disparate conclusions about where this selectivity emerges and whether it plays a major role in motion vision [8-13]. To address these questions, we examined activity in the neuropil thought to be responsible for visual motion detection, the medulla, of Drosophila melanogaster in response to a range of visual stimuli using two-photon calcium imaging. We confirmed that the input neurons of the medulla, the LMCs, are not responsible for light-on and light-off selectivity. We then examined the pan-neural response of medulla neurons and found prominent selectivity for light-on and light-off in layers of the medulla associated with two anatomically derived pathways (L1/L2 associated) [14, 15]. We next examined the activity of prominent interneurons within each pathway (Mi1 and Tm1) and found that these neurons have corresponding selectivity for light-on or light-off. These results provide direct evidence that motion is computed in parallel light-on and light-off pathways, demonstrate that this selectivity emerges in neurons immediately downstream of the LMCs, and specify where crucial elements of motion computation occur.
The Drosophila cerebrum originates from about 100 neuroblasts per hemisphere, with each neuroblast producing a characteristic set of neurons. Neurons from a neuroblast are often so diverse that many neuron types remain unexplored. We developed new genetic tools that target neuroblasts and their diverse descendants, increasing our ability to study fly brain structure and development. Common enhancer-based drivers label neurons on the basis of terminal identities rather than origins, which provides limited labeling in the heterogeneous neuronal lineages. We successfully converted conventional drivers that are temporarily expressed in neuroblasts, into drivers expressed in all subsequent neuroblast progeny. One technique involves immortalizing GAL4 expression in neuroblasts and their descendants. Another depends on loss of the GAL4 repressor, GAL80, from neuroblasts during early neurogenesis. Furthermore, we expanded the diversity of MARCM-based reagents and established another site-specific mitotic recombination system. Our transgenic tools can be combined to map individual neurons in specific lineages of various genotypes.
Despite the importance of the insect nervous system for functional and developmental neuroscience, descriptions of insect brains have suffered from a lack of uniform nomenclature. Ambiguous definitions of brain regions and fiber bundles have contributed to the variation of names used to describe the same structure. The lack of clearly determined neuropil boundaries has made it difficult to document precise locations of neuronal projections for connectomics study. To address such issues, a consortium of neurobiologists studying arthropod brains, the Insect Brain Name Working Group, has established the present hierarchical nomenclature system, using the brain of Drosophila melanogaster as the reference framework, while taking the brains of other taxa into careful consideration for maximum consistency and expandability. The following summarizes the consortium’s nomenclature system and highlights examples of existing ambiguities and remedies for them. This nomenclature is intended to serve as a standard of reference for the study of the brain of Drosophila and other insects.
The study of synaptic specificity and plasticity in the CNS is limited by the inability to efficiently visualize synapses in identified neurons using light microscopy. Here, we describe synaptic tagging with recombination (STaR), a method for labeling endogenous presynaptic and postsynaptic proteins in a cell-type-specific fashion. We modified genomic loci encoding synaptic proteins within bacterial artificial chromosomes such that these proteins, expressed at endogenous levels and with normal spatiotemporal patterns, were labeled in an inducible fashion in specific neurons through targeted expression of site-specific recombinases. Within the Drosophila visual system, the number and distribution of synapses correlate with electron microscopy studies. Using two different recombination systems, presynaptic and postsynaptic specializations of synaptic pairs can be colabeled. STaR also allows synapses within the CNS to be studied in live animals noninvasively. In principle, STaR can be adapted to the mammalian nervous system.