Random sample of points in a reconstructed neuron from zebrafinch neural tissue. Points are color coded with respect to the directionality of the neuron. Credit: Sheridan et al.
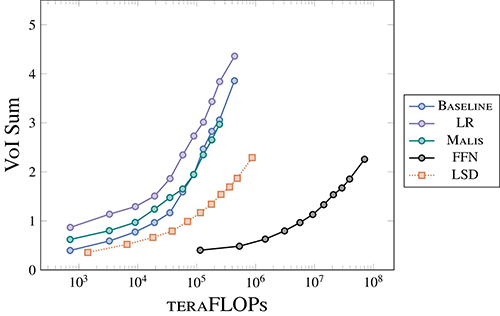
The new method, called Local Shape Descriptors (LSDs), allows researchers to analyze large electron microscopy data sets as accurately as state-of-the-art approaches but with computing power close to traditional neural networks. The freely available method, published in Nature Methods on December 30, 2022, is just as accurate and 100 times more efficient, enabling researchers to segment neurons using computer clusters found at research institutions worldwide.
“I see this contribution as a democratization of these kinds of algorithms,” says Janelia Group Leader Jan Funke.
Local shape descriptors
Electron microscopy has revolutionized the field of connectomics, allowing scientists to image the brain at nanometer scale and study connections between individual neurons. But these methods generate massive amounts of data, and the only way for scientists to efficiently analyze these images is by using algorithms run on huge clusters of computers. These state-of-the-art methods are very accurate but too computationally expensive for most researchers.

On the other hand, traditional methods of identifying individual cells are computationally efficient but lose accuracy when applied to large data sets. Traditional neural networks use information about cells’ boundaries to predict where they are in an image. These methods work well to identify simple-shaped cells in high-resolution microscopy images. But it can be a struggle for them to pick out individual neurons – long, thin shapes that span hundreds of microns – when they are jumbled up like a plate of spaghetti.
To overcome this, LSDs use information about the neuron in addition to its boundary, including its orientation and the thickness of its membrane. A network is then trained to identify these additional details in an image, making the new method better at pinpointing individual neurons than a traditional method.
“We force it to pay better attention to what is going on around the neuron to get a better estimate of whether there is a boundary or not,” says Funke.
 LSDs bridge the gap between traditional and state-of-the-art methods, says Sheridan, who is now in the Manor Lab at the Salk Institute. “It shows that you don’t need a technical, computationally complex method to achieve results on such a hard task,” he says. “If labs that have access to standard research infrastructure want to do connectomics, they can.”
LSDs bridge the gap between traditional and state-of-the-art methods, says Sheridan, who is now in the Manor Lab at the Salk Institute. “It shows that you don’t need a technical, computationally complex method to achieve results on such a hard task,” he says. “If labs that have access to standard research infrastructure want to do connectomics, they can.”
The method can also be used on other types of data. Julia Lyudchik, a PhD student in the Danzl Group at the Institute of Science and Technology Austria, used LSDs to analyze light microscopy images of mouse brain cells during a course taught by Funke and Sheridan at the Marine Biological Laboratory this past summer. It allowed Lyudchik to identify the neurons in her data in just five days, a task she said would likely have taken months on her own.
“This Local Shape Descriptors method has great potential,” Lyudchik says. “If you have 1 GPU on your laptop, you can already do a lot of things, which is very nice.”
###
Citation:
Arlo Sheridan, Tri M. Nguyen, Diptodip Deb, Wei-Chung Allen Lee, Stephan Saalfeld, Srinivas C. Turaga, Uri Manor and Jan Funke. “Local shape descriptors for neuron segmentation.” Nature Methods, Published December 30, 2022. DOI: 10.1038/s41592-022-01711-z
Media Contacts
Nanci Bompey




