Main Menu (Mobile)- Block
- Our Research
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Fly Facility
- Gene Targeting and Transgenics
- Immortalized Cell Line Culture
- Integrative Imaging
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing Software
- Scientific Computing Systems
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Fly Facility
- Gene Targeting and Transgenics
- Immortalized Cell Line Culture
- Integrative Imaging
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing Software
- Scientific Computing Systems
- Viral Tools
- Vivarium

The Cell Organelle Segmentation in Electron Microscopy Project Team uses machine learning and computer vision techniques to automatically identify and quantify all intracellular substructures within isotropic EM data.
Bridging from raw 3D EM data to more easily interpretable biological data.
The COSEM Project Team at Janelia Research campus uses computer vision and machine learning techniques to scalably detect subcellular structures, such as organelles, in datasets generated with next-generation volumetric electron microscopy.
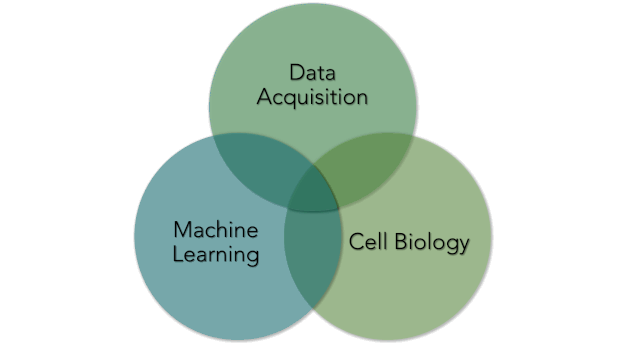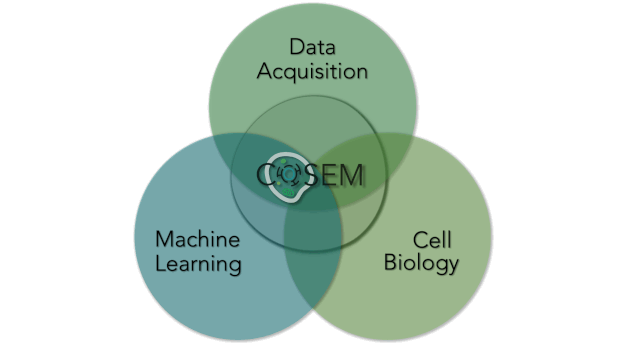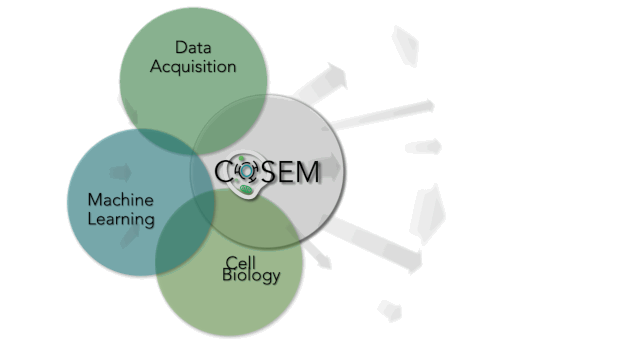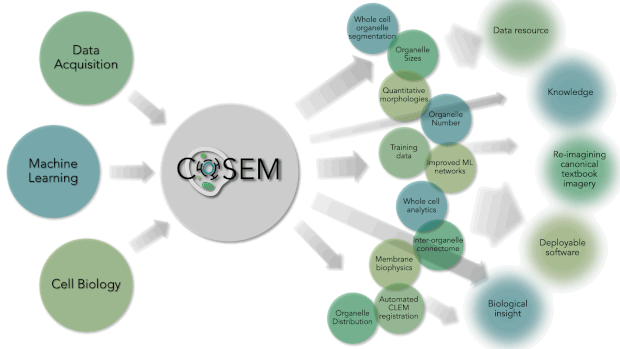
Within Janelia are some of the world’s leading experts in Machine Learning (Saalfeld lab and Funke lab), Cell Biology (Lippincott-Schwartz lab), and large-volume, high-resolution EM data acquisition (Hess lab and FIB-SEM Technology Group). At their intersection lies a unique area of research. COSEM is expanding that intersect and driving forward our tools and knowledge with respect to identifying subcellular structures within EM data in an automated way. As a result, the products that COSEM provides includes quantitative analytics and data resources. It will allow us to reimagine our canonical textbook image of the inside of a cell and provide biological insight and knowledge and do this in a way that is open and accessible to the broader scientific community and public.
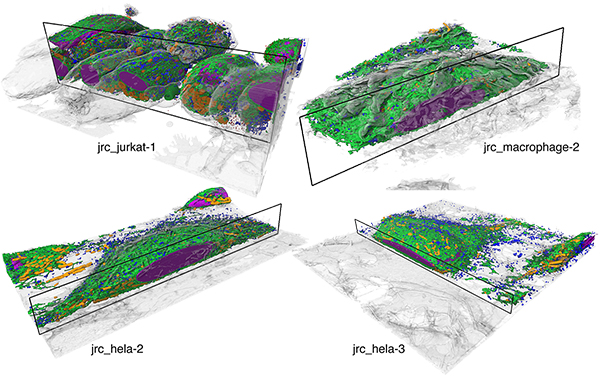 Our public repositories of raw datasets, training data, reconstructions, open source code, and models are an invitation to the scientific community to join our ongoing efforts to improve and further develop better and more robust automatic reconstruction and analysis methods for intra-cellular structures from FIB-SEM. We are optimistic that this will accelerate scientific discovery in both computational and biological domains leading to new and exciting insights and methods developments in the near future and beyond.
Our public repositories of raw datasets, training data, reconstructions, open source code, and models are an invitation to the scientific community to join our ongoing efforts to improve and further develop better and more robust automatic reconstruction and analysis methods for intra-cellular structures from FIB-SEM. We are optimistic that this will accelerate scientific discovery in both computational and biological domains leading to new and exciting insights and methods developments in the near future and beyond.
We invite you to visit our data portal OpenOrganelle to explore our data!
To find out more, please visit our Research page.
