The Saalfeld lab designs and develops methods and software for scalable automatic image analysis and collaborative manual and semi-automatic image annotation.
Lab Updates
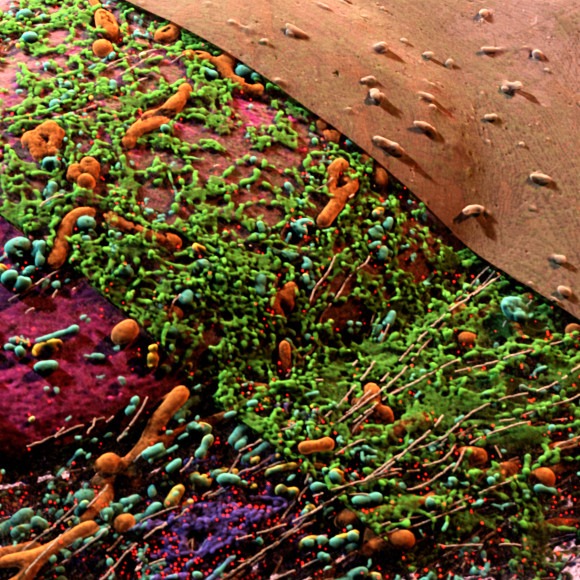
Cellular ultrastructure
All life on earth is composed of cells. Some organisms, like the
Cells, in single cell organisms, in collectives, or in highly specialized organ tissue implement an unbelievable variety of functions. They move, filter, digest and generate substrates, sense and send signals, ... all of this while they specialize and divide, to produce new cells, tidily following a genetically encoded plan, and maintaining the integrity of this genetic code.
Internally, all cells consist of organelles and other molecular structures such as the cytoskeleton and the components to maintain and transcribe and translate genetic information, many of which have been described in great detail based on studies using high resolution microscopy.
Cells are densely packed with these structures, but many of the mechanisms and interactions underlying their concerted function are poorly understood because it is hard to visualize them as a complete ensemble.
At Janelia, we have the unique opportunity to image whole cells and tissues at a resolution of a few nanometers per pixel with isotropic 3D electron microscopy using focused ion beam milling (FIB-SEM) developed by the lab of Harald Hess. At the same time, we can generate high spatial and temporal resolution movies of living cells and tissues with high resolution light microscopy developed by the labs of Eric Betzig and Philipp Keller. These impressive data demand that efficient automatic methods can be used to reliably and comprehensively reconstruct and analyze them. We develop these methods, at the forefront of machine learning, high dimensional computer vision, and big data analysis. We are an integral part of Janelia's CellMap project and collaborate with labs from Computation and Theory, Molecular Tools and Imaging, and the new research direction 4D Cellular Physiology to manage and process these impressive high dimensional microscopy data. We create tools to annotate and analyse these datasets, to provide ground truth data for training and validation of machine learning methods. We curate large public datasets to invite the scientific community to join our exploration, and we share all our tools and code as open source software.
Neural Connectivity Reconstruction at Scale
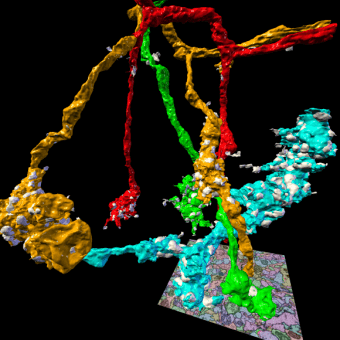
3D rendering of selected presynaptic neurons and synapses automatically reconstructed from serial section transmission electron microscopy of the Drosophila mushroom body calyx.
The brain of Drosophila melanogaster consists of ∼100,000 neurons and fits into a box of approximately 500×400×200 μm3. Individual neurons can span processes across distances of several 100 μm, their individual projections, however, can be as thin as 40 nm in diameter. Today, only Electron Microscopy (EM) offers a sufficient combination of spatial resolution, field of view, and throughput to image a biological nervous system of this size in its entirety and to potentially enable the reconstruction of the complete wiring diagram. At Janelia, we are using two complementary EM imaging techniques, (1) serial block-face scanning combined with ion beam milling (FIB-SEM, Hess lab, FlyEM project team) which generates isotropic image data at a resolution of typically 8 nm3 per voxel, and (2) serial section transmission imaging (ssTEM, Bock and Fetter labs) generating non-isotropic image data of typically 4×4×40 nm3 per voxel. At that resolution, a Drosophila brain fits into ∼40–80 trillion voxels.
Connectivity analysis from Electron Microscopy (EM) images requires both efficient automation and manual proofreading. Current state-of-the-art automatic image analysis methods have high demands on data quality to deliver satisfying results within reasonable time. Both FIB-SEM and ssTEM, however, come with their own set of preparation and imaging artifacts that make automatic and manual reconstruction a non-trivial endeavor. We are developing new automatic methods and user interfaces to compensate for these artifacts and to enable high quality automatic connectivity reconstruction from compromised data and complementary imaging modalities. In order to cope with the scale of the project, we pay significant attention to software design with focus on efficiency, scalability and re-usability.
