We are interested in developing and applying an experimental approach to neurobiology based on the comprehensive identification and manipulation of individual cell types and circuit components.
One of the great successes of biological research over the past three decades has been the use of genetic analysis to discover the pathways that control animal development. The success of this approach was largely dependent on three factors: (1) the genes themselves directly encode proteins, the primary functional units of these pathways; (2) nearly all the genes have been enumerated and described through analysis of genome sequences; and (3) powerful methods had been developed both to screen all the genes for their contribution to a given process and to inactivate, in a controlled way, the function of individual genes.
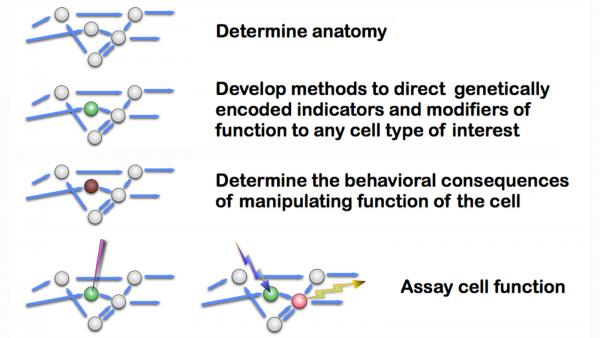
It is clear that the building blocks of the nervous system and individual neuronal circuits are not genes, but cells. And so the genetic methods that were so powerful in elucidating embryonic development will be of limited use in probing the function of the nervous system. Instead, we will need to be able to assay and manipulate the function of individual cells, if not the individual connections between cells, with the same facility as we can now manipulate genes. The diagram below shows the overall outline of this approach.
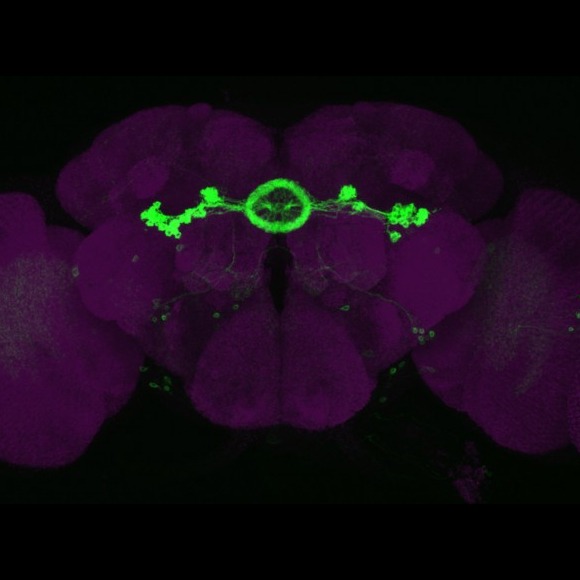
A variety of genetically encoded probes have been developed to allow experimenters to monitor and alter the activity of individual cells, and this remains an active and important area of research in many laboratories. The utility of these probes depends on the precision with which their expression can be directed to small subsets of cells in reproducible, controllable, and convenient ways. Providing the tools required to accomplish this task in the Drosophila nervous system of is a primary objective of our current research.
Another goal of our laboratory is to develop methods for light-level neuroanatomy and apply them in a comprehensive manner to key areas of the fly brain. Our current focus has been on the mushroom bodies, the central complex and the visual system. We work closely with others at Janelia performing neuroanatomy at the electron microscopic level in an effort to determine the precise wiring diagram—or connectome—of fly nervous system. In this way, we can develop the capability to image the activity and manipulate the function of specific nodes of the fly connectome.
Finally, we use the tools and methods we develop, usually in collaboration with other laboratories, to study the neuronal circuits underlying the function of specific brain areas involved in learning and memory, sleep regulation, visual perception and sensory integration.
Over the next decade, those of us working with Drosophila can expect to have comprehensive datasets and powerful tools with which to study the fly’s brain. These efforts should provide the first indication of whether the experimental and theoretical approaches envisioned by the BRAIN Initiative will be adequate to achieve “understanding” of how a nervous system generates sophisticated behaviors.
Current Collaborating Labs:
Michael Reiser
Gwyneth Card
Vivek Jayaram
Loren Looger
Larry Abbott
FlyLight Project Team
FlyEM Project Team
Glenn Turner (CSHL)
Richard Axel (Columbia)
Michael Nitabach (Yale)
Axel Borst (MPI, Martinsried)
Hirumo Tanimoto (Tohku Univ)
Roland Strauss (Mainz)