Organ function requires the synchronized regulation of numerous biological processes occurring across different scales to sense and adapt to changes in the external environment. Our lab will study how organelles within cells are regulated by tissue microenvironments and how they adapt to diurnal fluctuations in nutrient availability.
Investigating the Effects of Tissue Microenvironments Across Biological Scales
Biological Scales
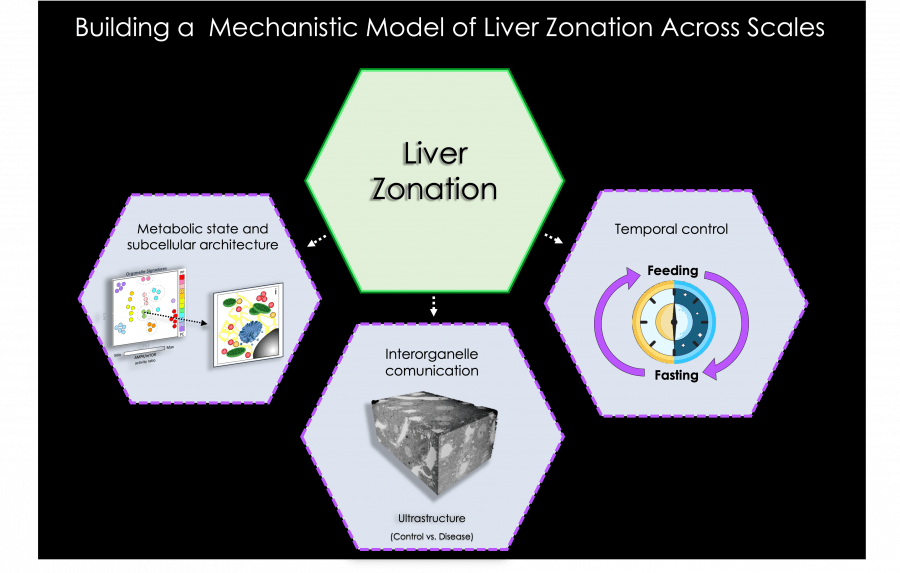
Metabolic flexibility in organs relies on fundamental biological processes coordinated within defined tissue architectures and modulated by external signals across biological scales. These external cues generate unique microenvironments and allow the segregation and regulation of distinct cellular tasks within metabolic organs. One such an organ is the liver, in which cells are organized into repeated hexagonal units known as liver lobules. These lobules possess heterogeneous populations of hepatocytes influenced by oxygen, nutrient, and hormone gradients formed by blood flowing from the portal to central site of the hepatic acinus. This unidirectional flow promotes different microenvironments giving rise to functionally distinct hepatocytes, a phenomenon known as "metabolic liver zonation". This segregation of specialized tasks not only promotes the compartmentalization of energetically demanding processes, but also makes hepatocytes susceptible to various pathologies in a zone-dependent manner. While liver zonation has resurfaced as a critical component shaping liver function and pathophysiology, the cues and mechanisms underlying liver zonation and its regulation at the tissue, cellular, and subcellular levels are much less understood.
To read more about liver zonation please see :
Thomas Kietzmann (2017), Shani Ben-Moshe and Shalev Itzkovitz (2019), and Rory P. Cunningham and Natalie Porat-Shliom (2021)
What determines the tissue microenvironment?
In addition to cues derived from blood circulation, paracrine signals coming from resident immune cells and molecules secreted from neurons innervating the organs have been suggested as factors regulating local microenvironments. Our lab, in collaboration with other labs at Janelia ( 4DCP ), will also explore the role of immune cells and neurons for the maintenance of microenvironments in the liver and the regulation of hepatocyte heterogeneity.
Implementing a tissue-to-organelle scale analysis approach:
At the subcellular level, organelles within hepatocytes conduct numerous metabolic tasks and respond to external signals (e.g., hormonal or nutritional cues), changing their morphology, numbers, and inter-organelle connections to meet the metabolic needs of the cell. Advances in fluorescence and ultrastructure microscopy in vitro have expanded our understanding on how different organelles undergo remodeling of their signatures and protein composition to adapt to various extracellular cues and changes in cellular bioenergetics. However, the intricate regulation of these organelle adaptive responses, in the context of physiological tissue microenvironments have not yet been revealed. This gap in knowledge stems from lack of readily available tools and multiscale analysis to simultaneously study multiple organelles and their dynamics within cells in the context of the intact tissue. This has limited our ability to identify zone-specific vulnerabilities that cause adverse outcomes affecting organelle dynamics, which could contribute to deregulation of metabolic flexibility and the onset of different disorders including liver pathologies.
The objective of our research program is to study metabolic liver zonation across scales to determine:
- How are organelles regulated within different zones ?
- How are organelle signatures modulated during feeding-fasting cycles ?
- How are organelles affected during liver illnesses ?
In addition, we are interested in how distinct metabolic states, arising from different microenvironments, influence the dynamics and nanoscale organization of different organelles within organs.
This will be achieved by implementing state-of-the-art imaging technologies ( light-sheet microscopy, intravital microscopy ( IVM ), multispectral unmixing, and ultrastructure microscopy ), new genetic tools ( e.g. TEMPO ) and collaborating with machine learning experts.