Main Menu (Mobile)- Block
- Overview
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
Our research lies at the intersection of AI and science, with a particular focus on neuroscience, biomechanics, optics, and protein engineering. We model the brain and the body in order to understand neural computation. We combine differentiable simulations of optical systems to develop a new kind of programmable microscope. We engineer new protein sensors using machine learning and mechanistic models of protein function.
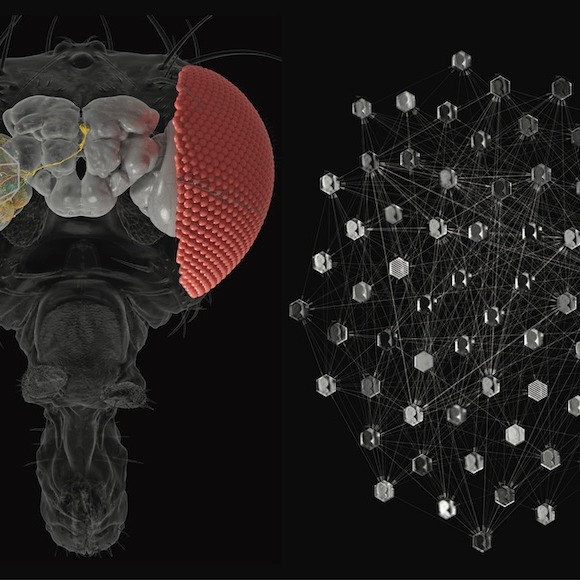
Recent technological advances in light and electron microscopy, optogenetics, single cell RNA sequencing, and more allow us to measure and perturb neural networks in unprecedented detail. These new data can be used understand how the structure of a neural circuit gives rise to its function -- how networks of neurons are connected, how cells are organized into cell types, and what activity and computation they produce. Our lab develops machine learning algorithms to map neural connectivity, and statistical models to characterize neural activity and to relate activity to connectivity. Projects currently underway include:
- Using deep neural networks and variational autoencoders to predict the spiking activity of neurons, infer their connectivity in vivo, and understand how neurons integrate their synaptic inputs.
- Building statistical models of neural activity and connectivity.
- Connectome driven computational models of neural circuits to understand their function.
- Discovering cell types and their basis, from single cell RNA sequencing data.
We have also been developing new computational microscopy techniques based on differentiable wave optical models of programmable microscopes, and deep neural networks. Projects in this space include:
- New methods for light field microscopy.
- Deep learning and variational autoencoder based algorithms for single molecule localization microscopy.
Finally, we have recently started building machine learning models for in silico protein engineering, starting with calcium indicators.
Check out some of our lab's collaborative work on GitHub.
