Expression Patterns of GAL4 and LexA Driver Lines
 Commonly referred to as "FlyLight External Website" or "FLEW", this website catalogs the imagery and associated data for Gen 1 GAL4 and LexA imagery From the Rubin and Truman Labs at Janelia Research Campus, as well as the Mann (Columbia) and Doe (Oregon) Labs. Users may search by genetic line, anatomical expression (adult and larval), and anatomy (larval and imaginal disc).
Commonly referred to as "FlyLight External Website" or "FLEW", this website catalogs the imagery and associated data for Gen 1 GAL4 and LexA imagery From the Rubin and Truman Labs at Janelia Research Campus, as well as the Mann (Columbia) and Doe (Oregon) Labs. Users may search by genetic line, anatomical expression (adult and larval), and anatomy (larval and imaginal disc).
FlyLight Split-GAL4 Driver Collection
 This website catalogs the imagery and associated data for Split-GAL4 imagery from the Rubin Lab at Janelia Research Campus. Users may search by genetic line, anatomical expression, or fragment (AD/DBD). The current version of this website covers Mushroom Body lines as published in "The neuronal architecture of the mushroom body provides a logic for associative learning" (Aso, et al.).
This website catalogs the imagery and associated data for Split-GAL4 imagery from the Rubin Lab at Janelia Research Campus. Users may search by genetic line, anatomical expression, or fragment (AD/DBD). The current version of this website covers Mushroom Body lines as published in "The neuronal architecture of the mushroom body provides a logic for associative learning" (Aso, et al.).
Janelia Workstation
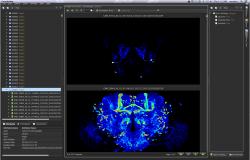 The Janelia Workstation is a discovery platform used to analyze and annotate imagery for individual projects, while assembling shared data resources such as neuron atlases. It was originally applied to Drosophila neuronal anatomy and neuroblast lineage analysis and is currently being extended to support mouse whole-brain projection tracing. The Workstation is both a pipeline management system that extracts entities from 3D imagery and a suite of tools that enable scientists to annotate large imagery datasets. The kernel of the design is an Entity-Attribute-Value (EAV) graph permitting any element to be annotated by users with custom ontologies. Currently there are ~30 million entities in the graph, with ~700,000 manual annotations. By allowing any entity to have multiple parents, the graph can be re-arranged by each user to suit their research needs without copying the underlying image data.
The Janelia Workstation is a discovery platform used to analyze and annotate imagery for individual projects, while assembling shared data resources such as neuron atlases. It was originally applied to Drosophila neuronal anatomy and neuroblast lineage analysis and is currently being extended to support mouse whole-brain projection tracing. The Workstation is both a pipeline management system that extracts entities from 3D imagery and a suite of tools that enable scientists to annotate large imagery datasets. The kernel of the design is an Entity-Attribute-Value (EAV) graph permitting any element to be annotated by users with custom ontologies. Currently there are ~30 million entities in the graph, with ~700,000 manual annotations. By allowing any entity to have multiple parents, the graph can be re-arranged by each user to suit their research needs without copying the underlying image data.
Architecturally, the Workstation is implemented as an n-tier application with client, application, database, and cluster components. The client is used as a hub for launching spoke tools, such as Fiji, Vaa3d, and a custom application for manually annotating neurons called Neuron Annotator. Technologies used include JBoss, WebDAV, MySQL, Solr, and a 10,000 node CPU cluster.
Split Picker
 The Split Picker is a web application that provides a side-by-side view of images for a reference line with images for a series of potential targets. Potential crosses may be selected, and both projections and LSMs can be downloaded. Although currently optimized for the Dickson Lab, this application will allow for viewing and split selection for any GAL4 and LexA screening lines.
The Split Picker is a web application that provides a side-by-side view of images for a reference line with images for a series of potential targets. Potential crosses may be selected, and both projections and LSMs can be downloaded. Although currently optimized for the Dickson Lab, this application will allow for viewing and split selection for any GAL4 and LexA screening lines.
Transmogrifier
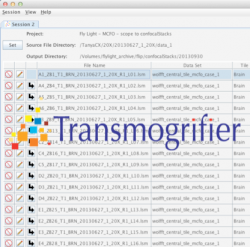 The Janelia Transmogrifier (tmog) supports both the collection of meta data for files and the reliable, tracked transfer of files from source systems to centralized storage. Numerous plug-ins and configurations have been developed to integrate with remote databases and systems for validating and storing collected meta data.
The Janelia Transmogrifier (tmog) supports both the collection of meta data for files and the reliable, tracked transfer of files from source systems to centralized storage. Numerous plug-ins and configurations have been developed to integrate with remote databases and systems for validating and storing collected meta data.