Main Menu (Mobile)- Block
- Overview
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
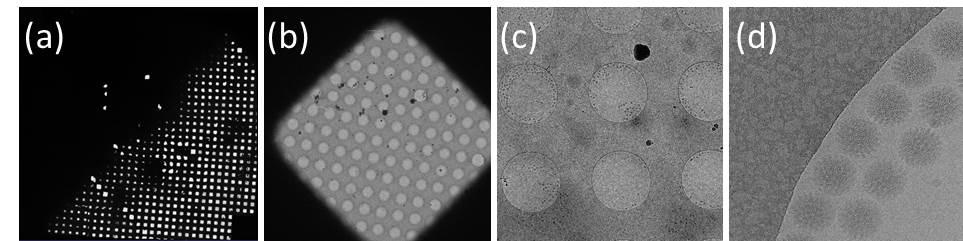
A single particle analysis example from Janelia Krios2
Single particle data collection typically collects one exposure/movie stack at one sample area and moves to a new sample area for the next exposure/movie stack. It involves imaging a cryo grid at different scales/magnifications: (a) overview of an entire cryoEM grid. An ice thickness gradient is clearly visible. The ice is too thick on the top left half of the grid and becomes thinner and thinner going bottom right where many grid squares separated by grid bars can be seen. (b) image of one grid square. There is a carbon film in the grid square with patterned holes over which vitrified ice with biological samples is suspended. (c) a medium magnification image to show a few holes in a grid square. (d) The final collection data from a small portion of one hole in (c) at high magnification. Bovine rotavirus particles can be seen in vitrified ice along the edge of the hole. Data collected on Krios2 by Tim Grant in Niko Grigorieff’s lab at Janelia.
Three dimensional (3D) reconstruction of bovine rotavirus from a data set of 4000 particles collected on Janelia Krios2 (Tim Grant and Niko Grigorieff).
Capsid protein VP6 monomer at 2.6 angstrom resolution from the 3D reconstruction on bovine rotavirus (Tim Grant and Niko Grigorieff). (A) density map of an entire VP6 monomer with atomic model; (B) Zoomed in region shows the side chain density; (C) zoomed in region shows that water molecules are resolved.