Filter
Associated Lab
Publication Date
- 2014 (1) Apply 2014 filter
- 2012 (1) Apply 2012 filter
- 2011 (1) Apply 2011 filter
- 2010 (2) Apply 2010 filter
- 2009 (1) Apply 2009 filter
- 2008 (2) Apply 2008 filter
- 2002 (1) Apply 2002 filter
- 2000 (2) Apply 2000 filter
- 1999 (1) Apply 1999 filter
- 1997 (1) Apply 1997 filter
- 1995 (2) Apply 1995 filter
- 1994 (2) Apply 1994 filter
- 1993 (2) Apply 1993 filter
- 1992 (1) Apply 1992 filter
- 1991 (2) Apply 1991 filter
- 1990 (3) Apply 1990 filter
- 1989 (2) Apply 1989 filter
- 1987 (2) Apply 1987 filter
- 1986 (1) Apply 1986 filter
- 1985 (1) Apply 1985 filter
- 1984 (1) Apply 1984 filter
- 1983 (1) Apply 1983 filter
- 1982 (2) Apply 1982 filter
- 1981 (1) Apply 1981 filter
- 1979 (1) Apply 1979 filter
- 1973 (1) Apply 1973 filter
Type of Publication
- Remove Non-Janelia filter Non-Janelia
38 Publications
Showing 1-10 of 38 resultsFlexible goal-driven orientation requires that the position of a target be stored, especially in case the target moves out of sight. The capability to retain, recall and integrate such positional information into guiding behaviour has been summarized under the term spatial working memory. This kind of memory contains specific details of the presence that are not necessarily part of a long-term memory. Neurophysiological studies in primates indicate that sustained activity of neurons encodes the sensory information even though the object is no longer present. Furthermore they suggest that dopamine transmits the respective input to the prefrontal cortex, and simultaneous suppression by GABA spatially restricts this neuronal activity. Here we show that Drosophila melanogaster possesses a similar spatial memory during locomotion. Using a new detour setup, we show that flies can remember the position of an object for several seconds after it has been removed from their environment. In this setup, flies are temporarily lured away from the direction towards their hidden target, yet they are thereafter able to aim for their former target. Furthermore, we find that the GABAergic (stainable with antibodies against GABA) ring neurons of the ellipsoid body in the central brain are necessary and their plasticity is sufficient for a functional spatial orientation memory in flies. We also find that the protein kinase S6KII (ignorant) is required in a distinct subset of ring neurons to display this memory. Conditional expression of S6KII in these neurons only in adults can restore the loss of the orientation memory of the ignorant mutant. The S6KII signalling pathway therefore seems to be acutely required in the ring neurons for spatial orientation memory in flies.
The rhodopsin genes of Drosophila melanogaster are expressed in nonoverlapping subsets of photoreceptor cells within the insect visual system. Two of these genes, Rh3 and Rh4, are known to display complementary expression patterns in the UV-sensitive R7 photoreceptor cell population of the compound eye. In addition, we find that Rh3 is expressed in a small group of paired R7 and R8 photoreceptor cells at the dorsal eye margin that are apparently specialized for the detection of polarized light. In this paper we present a detailed characterization of the cis-acting requirements of both Rh3 and Rh4. Promoter deletion series demonstrate that small regulatory regions (less than 300 bp) of both R7 opsin genes contain DNA sequences sufficient to generate their respective expression patterns. Individual cis-acting elements were further identified by oligonucleotide-directed mutagenesis guided by interspecific sequence comparisons. Our results suggest that the Drosophila rhodopsin genes share a simple bipartite promoter structure, whereby the proximal region constitutes a functionally equivalent promoter "core" and the distal region determines cell-type specificity. The expression patterns of several hybrid rhodopsin promoters, in which all or part of the putative core regions have been replaced with the analogous regions of different rhodopsin promoters, provide additional evidence in support of this model.
We have constructed a series of strains to facilitate the generation and analysis of clones of genetically distinct cells in developing and adult tissues of Drosophila. Each of these strains carries an FRT element, the target for the yeast FLP recombinase, near the base of a major chromosome arm, as well as a gratuitous cell-autonomous marker. Novel markers that carry epitope tags and that are localized to either the cell nucleus or cell membrane have been generated. As a demonstration of how these strains can be used to study a particular gene, we have analyzed the developmental role of the Drosophila EGF receptor homolog. Moreover, we have shown that these strains can be utilized to identify new mutations in mosaic animals in an efficient and unbiased way, thereby providing an unprecedented opportunity to perform systematic genetic screens for mutations affecting many biological processes.
We have made a P-element derivative called Pc[ry], which carries the selectable marker gene rosy, but which acts like a nondefective, intact P element. It transposes autonomously into the germline chromosomes of an M-strain Drosophila embryo and it mobilizes in trans the defective P elements of the singed-weak allele. Frameshift mutations introduced into any of the four major open reading frames of the P sequence were each sufficient to eliminate the transposase activity, but none affected signals required in cis for transposition of the element. Complementation tests between pairs of mutant elements suggest that a single polypeptide comprises the transposase. We have examined transcripts of P elements both from natural P strains and from lines containing only nondefective Pc[ry] elements, and have identified two RNA species that appear to be specific for autonomous elements.
To compare appetitive and aversive visual memories of the fruit fly Drosophila melanogaster, we developed a new paradigm for classical conditioning. Adult flies are trained en masse to differentially associate one of two visual conditioned stimuli (CS) (blue and green light as CS) with an appetitive or aversive chemical substance (unconditioned stimulus or US). In a test phase, flies are given a choice between the paired and the unpaired visual stimuli. Associative memory is measured based on altered visual preference in the test. If a group of flies has, for example, received a sugar reward with green light in the training, they show a significantly higher preference for the green stimulus during the test than another group of flies having received the same reward with blue light. We demonstrate critical parameters for the formation of visual appetitive memory, such as training repetition, order of reinforcement, starvation, and individual conditioning. Furthermore, we show that formic acid can act as an aversive chemical reinforcer, yielding weak, yet significant, aversive memory. These results provide a basis for future investigations into the cellular and molecular mechanisms underlying visual memory and perception in Drosophila.
We tested whether Drosophila larvae can associate odours with a mechanosensory disturbance as a punishment, using substrate vibration conveyed by a loudspeaker (buzz:). One odour (A) was presented with the buzz, while another odour (B) was presented without the buzz (A/B training). Then, animals were offered the choice between A and B. After reciprocal training (A/B), a second experimental group was tested in the same way. We found that larvae show conditioned escape from the previously punished odour. We further report an increase of associative performance scores with the number of punishments, and an increase according to the number of training cycles. Within the range tested (between 50 and 200 Hz), however, the pitch of the buzz does not apparently impact associative success. Last, but not least, we characterized odour-buzz memories with regard to the conditions under which they are behaviourally expressed--or not. In accordance with what has previously been found for associative learning between odours and bad taste (such as high concentration salt or quinine), we report that conditioned escape after odour-buzz learning is disabled if escape is not warranted, i.e. if no punishment to escape from is present during testing. Together with the already established paradigms for the association of odour and bad taste, the present assay offers the prospect of analysing how a relatively simple brain orchestrates memory and behaviour with regard to different kinds of 'bad' events.
Cellular ultrastructures for signal integration are unknown in any nervous system. The ellipsoid body (EB) of the Drosophila brain is thought to control locomotion upon integration of various modalities of sensory signals with the animal internal status. However, the expected excitatory and inhibitory input convergence that virtually all brain centres exhibit is not yet described in the EB. Based on the EB expression domains of genetic constructs from the choline acetyl transferase (Cha), glutamic acid decarboxylase (GAD) and tyrosine hydroxylase (TH) genes, we identified a new set of neurons with the characteristic ring-shaped morphology (R neurons) which are presumably cholinergic, in addition to the existing GABA-expressing neurons. The R1 morphological subtype is represented in the Cha- and TH-expressing classes. In addition, using transmission electron microscopy, we identified a novel type of synapse in the EB, which exhibits the precise array of two independent active zones over the same postsynaptic dendritic domain, that we named 'agora'. This array is compatible with a coincidence detector role, and represents ~8% of all EB synapses in Drosophila. Presumably excitatory R neurons contribute to coincident synapses. Functional silencing of EB neurons by driving genetically tetanus toxin expression either reduces walking speed or alters movement orientation depending on the targeted R neuron subset, thus revealing functional specialisations in the EB for locomotion control.
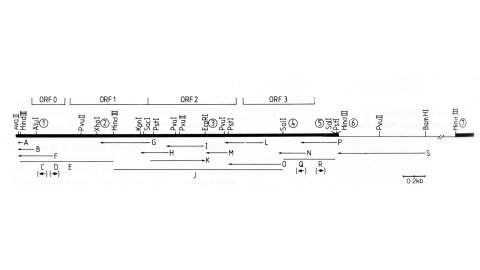
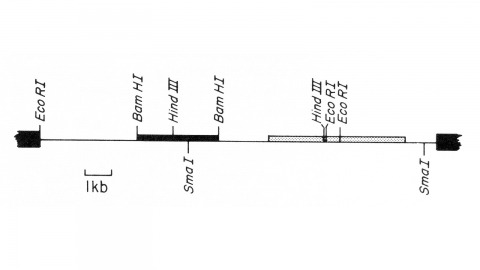
We describe the isolation of a cloned DNA segment carrying unique sequences from the white locus of Drosophila melanogaster. Sequences within the cloned segment are shown to hybridize in situ to the white locus region on the polytene chromosomes of both wild-type strains and strains carrying chromosomal rearrangements whose breakpoints bracket the white locus. We further show that two small deficiency mutations, deleting white locus genetic elements but not those of complementation groups contiguous to white, delete the genomic sequences corresponding to a portion of the cloned segment. The strategy we have employed to isolate this cloned segment exploits the existence of an allele at the white locus containing a copy of a previously cloned transposable, reiterated DNA sequence element. We describe a simple, rapid method for retrieving cloned segments carrying a copy of the transposable element together with contiguous sequences corresponding to this allele. The strategy described is potentially general and we discuss its application to the cloning of the DNA sequences of other genes in Drosophila, including those identified only by genetic analysis and for which no RNA product is known.
A comparative analysis of the genomes of Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae-and the proteins they are predicted to encode-was undertaken in the context of cellular, developmental, and evolutionary processes. The nonredundant protein sets of flies and worms are similar in size and are only twice that of yeast, but different gene families are expanded in each genome, and the multidomain proteins and signaling pathways of the fly and worm are far more complex than those of yeast. The fly has orthologs to 177 of the 289 human disease genes examined and provides the foundation for rapid analysis of some of the basic processes involved in human disease.
Apoptotic cell death is a mechanism by which organisms eliminate superfluous or harmful cells. Expression of the cell death regulatory protein REAPER (RPR) in the developing Drosophila eye results in a small eye owing to excess cell death. We show that mutations in thread (th) are dominant enhancers of RPR-induced cell death and that th encodes a protein homologous to baculovirus inhibitors of apoptosis (IAPs), which we call Drosophila IAP1 (DIAP1). Overexpression of DIAP1 or a related protein, DIAP2, in the eye suppresses normally occurring cell death as well as death due to overexpression of rpr or head involution defective. IAP death-preventing activity localizes to the N-terminal baculovirus IAP repeats, a motif found in both viral and cellular proteins associated with death prevention.