Filter
Associated Lab
- Ahrens Lab (3) Apply Ahrens Lab filter
- Aso Lab (6) Apply Aso Lab filter
- Baker Lab (2) Apply Baker Lab filter
- Betzig Lab (11) Apply Betzig Lab filter
- Branson Lab (6) Apply Branson Lab filter
- Cardona Lab (5) Apply Cardona Lab filter
- Chklovskii Lab (2) Apply Chklovskii Lab filter
- Cui Lab (5) Apply Cui Lab filter
- Darshan Lab (1) Apply Darshan Lab filter
- Dickson Lab (2) Apply Dickson Lab filter
- Druckmann Lab (3) Apply Druckmann Lab filter
- Dudman Lab (3) Apply Dudman Lab filter
- Eddy/Rivas Lab (4) Apply Eddy/Rivas Lab filter
- Egnor Lab (1) Apply Egnor Lab filter
- Fetter Lab (5) Apply Fetter Lab filter
- Fitzgerald Lab (3) Apply Fitzgerald Lab filter
- Freeman Lab (7) Apply Freeman Lab filter
- Funke Lab (1) Apply Funke Lab filter
- Gonen Lab (5) Apply Gonen Lab filter
- Grigorieff Lab (8) Apply Grigorieff Lab filter
- Harris Lab (7) Apply Harris Lab filter
- Heberlein Lab (2) Apply Heberlein Lab filter
- Hess Lab (7) Apply Hess Lab filter
- Jayaraman Lab (4) Apply Jayaraman Lab filter
- Ji Lab (4) Apply Ji Lab filter
- Karpova Lab (1) Apply Karpova Lab filter
- Keleman Lab (2) Apply Keleman Lab filter
- Keller Lab (8) Apply Keller Lab filter
- Lavis Lab (5) Apply Lavis Lab filter
- Leonardo Lab (2) Apply Leonardo Lab filter
- Li Lab (3) Apply Li Lab filter
- Lippincott-Schwartz Lab (4) Apply Lippincott-Schwartz Lab filter
- Liu (Zhe) Lab (4) Apply Liu (Zhe) Lab filter
- Looger Lab (9) Apply Looger Lab filter
- Magee Lab (5) Apply Magee Lab filter
- Menon Lab (2) Apply Menon Lab filter
- Murphy Lab (1) Apply Murphy Lab filter
- Otopalik Lab (2) Apply Otopalik Lab filter
- Pachitariu Lab (2) Apply Pachitariu Lab filter
- Pastalkova Lab (3) Apply Pastalkova Lab filter
- Reiser Lab (2) Apply Reiser Lab filter
- Romani Lab (4) Apply Romani Lab filter
- Rubin Lab (16) Apply Rubin Lab filter
- Saalfeld Lab (4) Apply Saalfeld Lab filter
- Scheffer Lab (2) Apply Scheffer Lab filter
- Schreiter Lab (4) Apply Schreiter Lab filter
- Sgro Lab (3) Apply Sgro Lab filter
- Shroff Lab (1) Apply Shroff Lab filter
- Simpson Lab (4) Apply Simpson Lab filter
- Singer Lab (16) Apply Singer Lab filter
- Spruston Lab (7) Apply Spruston Lab filter
- Stern Lab (4) Apply Stern Lab filter
- Sternson Lab (7) Apply Sternson Lab filter
- Svoboda Lab (9) Apply Svoboda Lab filter
- Tebo Lab (2) Apply Tebo Lab filter
- Tillberg Lab (1) Apply Tillberg Lab filter
- Tjian Lab (6) Apply Tjian Lab filter
- Truman Lab (6) Apply Truman Lab filter
- Turaga Lab (2) Apply Turaga Lab filter
- Turner Lab (3) Apply Turner Lab filter
- Wang (Shaohe) Lab (1) Apply Wang (Shaohe) Lab filter
- Wu Lab (1) Apply Wu Lab filter
- Zlatic Lab (4) Apply Zlatic Lab filter
- Zuker Lab (2) Apply Zuker Lab filter
Associated Project Team
Publication Date
- December 2015 (15) Apply December 2015 filter
- November 2015 (22) Apply November 2015 filter
- October 2015 (18) Apply October 2015 filter
- September 2015 (20) Apply September 2015 filter
- August 2015 (17) Apply August 2015 filter
- July 2015 (18) Apply July 2015 filter
- June 2015 (20) Apply June 2015 filter
- May 2015 (20) Apply May 2015 filter
- April 2015 (24) Apply April 2015 filter
- March 2015 (21) Apply March 2015 filter
- February 2015 (36) Apply February 2015 filter
- January 2015 (21) Apply January 2015 filter
- Remove 2015 filter 2015
Type of Publication
252 Publications
Showing 241-250 of 252 resultsHuman memory stores vast amounts of information. Yet recalling this information is often challenging when specific cues are lacking. Here we consider an associative model of retrieval where each recalled item triggers the recall of the next item based on the similarity between their long-term neuronal representations. The model predicts that different items stored in memory have different probability to be recalled depending on the size of their representation. Moreover, items with high recall probability tend to be recalled earlier and suppress other items. We performed an analysis of a large data set on free recall and found a highly specific pattern of statistical dependencies predicted by the model, in particular negative correlations between the number of words recalled and their average recall probability. Taken together, experimental and modeling results presented here reveal complex interactions between memory items during recall that severely constrain recall capacity.
The mammalian taste system is responsible for sensing and responding to the five basic taste qualities: sweet, sour, bitter, salty and umami. Previously, we showed that each taste is detected by dedicated taste receptor cells (TRCs) on the tongue and palate epithelium. To understand how TRCs transmit information to higher neural centres, we examined the tuning properties of large ensembles of neurons in the first neural station of the gustatory system. Here, we generated and characterized a collection of transgenic mice expressing a genetically encoded calcium indicator in central and peripheral neurons, and used a gradient refractive index microendoscope combined with high-resolution two-photon microscopy to image taste responses from ganglion neurons buried deep at the base of the brain. Our results reveal fine selectivity in the taste preference of ganglion neurons; demonstrate a strong match between TRCs in the tongue and the principal neural afferents relaying taste information to the brain; and expose the highly specific transfer of taste information between taste cells and the central nervous system.
The crustacean stomatogastric ganglion (STG) receives descending neuromodulatory inputs from three anterior ganglia: the paired commissural ganglia (CoGs), and the single esophageal ganglion (OG). In this paper, we provide the first detailed and quantitative analyses of the short- and long-term effects of removal of these descending inputs (decentralization) on the pyloric rhythm of the STG. Thirty minutes after decentralization, the mean frequency of the pyloric rhythm dropped from 1.20 Hz in control to 0.52 Hz. Whereas the relative phase of pyloric neuron activity was approximately constant across frequency in the controls, after decentralization this changed markedly. Nine control preparations kept for 5–6 d in vitro maintained pyloric rhythm frequencies close to their initial values. Nineteen decentralized preparations kept for 5–6 d dropped slightly in frequency from those seen at 30 min following decentralization, but then displayed stable activity over 6 d. Bouts of higher frequency activity were intermittently seen in both control and decentralized preparations, but the bouts began earlier and were more frequent in the decentralized preparations. Although the bouts may indicate that the removal of the modulatory inputs triggered changes in neuronal excitability, these changes did not produce obvious long-lasting changes in the frequency of the decentralized preparations.
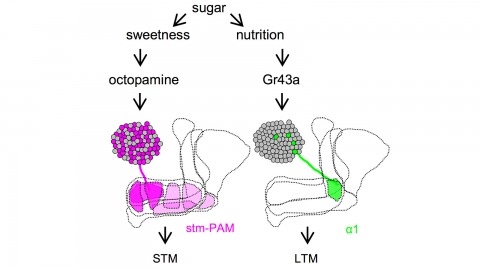
Drosophila melanogaster can acquire a stable appetitive olfactory memory when the presentation of a sugar reward and an odor are paired. However, the neuronal mechanisms by which a single training induces long-term memory are poorly understood. Here we show that two distinct subsets of dopamine neurons in the fly brain signal reward for short-term (STM) and long-term memories (LTM). One subset induces memory that decays within several hours, whereas the other induces memory that gradually develops after training. They convey reward signals to spatially segregated synaptic domains of the mushroom body (MB), a potential site for convergence. Furthermore, we identified a single type of dopamine neuron that conveys the reward signal to restricted subdomains of the mushroom body lobes and induces long-term memory. Constant appetitive memory retention after a single training session thus comprises two memory components triggered by distinct dopamine neurons.
Understanding how brain activation mediates behaviors is a central goal of systems neuroscience. Here, we apply an automated method for mapping brain activation in the mouse in order to probe how sex-specific social behaviors are represented in the male brain. Our method uses the immediate-early-gene c-fos, a marker of neuronal activation, visualized by serial two-photon tomography: the c-fos-GFP+ neurons are computationally detected, their distribution is registered to a reference brain and a brain atlas, and their numbers are analyzed by statistical tests. Our results reveal distinct and shared female and male interaction-evoked patterns of male brain activation representing sex discrimination and social recognition. We also identify brain regions whose degree of activity correlates to specific features of social behaviors and estimate the total numbers and the densities of activated neurons per brain areas. Our study opens the door to automated screening of behavior-evoked brain activation in the mouse.
Complex animal behaviors are built from dynamical relationships between sensory inputs, neuronal activity, and motor outputs in patterns with strategic value. Connecting these patterns illuminates how nervous systems compute behavior. Here, we study Drosophila larva navigation up temperature gradients toward preferred temperatures (positive thermotaxis). By tracking the movements of animals responding to fixed spatial temperature gradients or random temperature fluctuations, we calculate the sensitivity and dynamics of the conversion of thermosensory inputs into motor responses. We discover three thermosensory neurons in each dorsal organ ganglion (DOG) that are required for positive thermotaxis. Random optogenetic stimulation of the DOG thermosensory neurons evokes behavioral patterns that mimic the response to temperature variations. In vivo calcium and voltage imaging reveals that the DOG thermosensory neurons exhibit activity patterns with sensitivity and dynamics matched to the behavioral response. Temporal processing of temperature variations carried out by the DOG thermosensory neurons emerges in distinct motor responses during thermotaxis.
Three-helix bundles and coiled-coil motifs are well-established de novo designed scaffolds that have been investigated for their metal-binding and catalytic properties. Satisfaction of the primary coordination sphere for a given metal is sufficient to introduce catalytic activity and a given structure may catalyze different reactions dependent on the identity of the incorporated metal. Here we describe recent contributions in the de novo design of metalloenzymes based on three-helix bundles and coiled-coil motifs, focusing on non-heme systems for hydrolytic and redox chemistry.
The formation of functional neuronal circuits relies on accurate migration and proper axonal outgrowth of neuronal precursors. On the route to their targets migrating cells and growing axons depend on both, directional information from neurotropic cues and adhesive interactions mediated via extracellular matrix molecules or neighbouring cells. The inactivation of guidance cues or the interference with cell adhesion can cause severe defects in neuronal migration and axon guidance. In this study we have analyzed the function of the MAM domain containing glycosylphosphatidylinositol anchor 2A (MDGA2A) protein in zebrafish cranial motoneuron development. MDGA2A is prominently expressed in distinct clusters of cranial motoneurons, especially in the ones of the trigeminal and facial nerves. Analyses of MDGA2A knockdown embryos by light sheet and confocal microscopy revealed impaired migration and aberrant axonal outgrowth of these neurons; suggesting that adhesive interactions mediated by MDGA2A are required for the proper arrangement and outgrowth of cranial motoneuron subtypes.
Tsetse flies (Glossina spp.), vectors of African trypanosomes, are distinguished by their specialized reproductive biology, defined by adenotrophic viviparity (maternal nourishment of progeny by glandular secretions followed by live birth). This trait has evolved infrequently among insects and requires unique reproductive mechanisms. A key event in Glossina reproduction involves the transition between periods of lactation and nonlactation (dry periods). Increased lipolysis, nutrient transfer to the milk gland, and milk-specific protein production characterize lactation, which terminates at the birth of the progeny and is followed by a period of involution. The dry stage coincides with embryogenesis of the progeny, during which lipid reserves accumulate in preparation for the next round of lactation. The obligate bacterial symbiont Wigglesworthia glossinidia is critical to tsetse reproduction and likely provides B vitamins required for metabolic processes underlying lactation and/or progeny development. Here we describe findings that utilized transcriptomics, physiological assays, and RNA interference-based functional analysis to understand different components of adenotrophic viviparity in tsetse flies.
NAD(+) and NADH play crucial roles in a variety of biological processes including energy metabolism, mitochondrial functions, and gene expression. Multiple studies have indicated that NAD(+) administration can profoundly decrease oxidative cell death as well as ischemic and traumatic brain injury, suggesting NAD(+) metabolism as a promising therapeutic target for cerebral ischemia and head injury. Cumulating evidence has suggested that NAD(+) can produce its protective effects by multiple mechanisms, including preventing mitochondrial alterations, enhancing energy metabolism, preventing virtually all forms of cell death including apoptosis, necrosis and autophagy, inhibiting inflammation, directly increasing antioxidation capacity of cells and tissues, and activating SIRT1. Increasing evidence has also suggested that NADH metabolism is a potential therapeutic target for treating several neurological disorders. A number of studies have further indicated that multiple NAD(+)-dependent enzymes such as sirtuins, polymerase(ADP-ribose) polymerases (PARPs) and CD38 mediate cell death and multiple biological processes. In this article, an overview of the recent findings regarding the roles of NAD(+)/NADH and NAD(+)-dependent enzymes in cell death and ischemic brain injury is provided. These findings have collectively indicated that NAD(+)/NADH and NAD(+)-dependent enzymes play fundamental roles in oxidative stress-induced cell death and ischemic brain injury, which may become promising therapeutic targets for brain ischemia and multiple other neurological disorders.