Filter
Associated Lab
- Dudman Lab (1) Apply Dudman Lab filter
- Hermundstad Lab (1) Apply Hermundstad Lab filter
- Lavis Lab (1) Apply Lavis Lab filter
- Lee (Albert) Lab (1) Apply Lee (Albert) Lab filter
- Looger Lab (2) Apply Looger Lab filter
- Pachitariu Lab (1) Apply Pachitariu Lab filter
- Saalfeld Lab (2) Apply Saalfeld Lab filter
- Scheffer Lab (1) Apply Scheffer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Remove Sternson Lab filter Sternson Lab
- Svoboda Lab (4) Apply Svoboda Lab filter
- Tillberg Lab (3) Apply Tillberg Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
Associated Project Team
Publication Date
- 2023 (2) Apply 2023 filter
- 2022 (3) Apply 2022 filter
- 2021 (3) Apply 2021 filter
- 2020 (4) Apply 2020 filter
- 2019 (2) Apply 2019 filter
- 2017 (2) Apply 2017 filter
- 2016 (3) Apply 2016 filter
- 2015 (7) Apply 2015 filter
- 2014 (5) Apply 2014 filter
- 2013 (3) Apply 2013 filter
- 2012 (4) Apply 2012 filter
- 2011 (6) Apply 2011 filter
- 2010 (1) Apply 2010 filter
- 2009 (2) Apply 2009 filter
- 2008 (1) Apply 2008 filter
- 2005 (1) Apply 2005 filter
- 2004 (1) Apply 2004 filter
- 2002 (1) Apply 2002 filter
- 2001 (2) Apply 2001 filter
- 1998 (1) Apply 1998 filter
Type of Publication
54 Publications
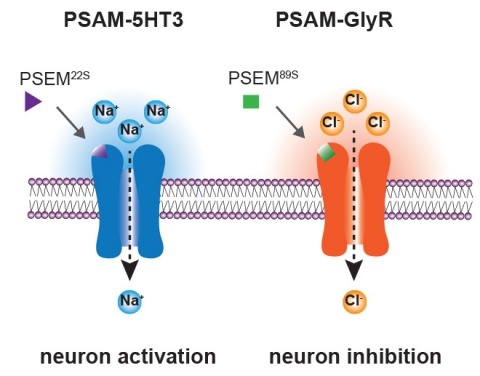
Showing 41-50 of 54 resultsIonic flux mediates essential physiological and behavioral functions in defined cell populations. Cell type-specific activators of diverse ionic conductances are needed for probing these effects. We combined chemistry and protein engineering to enable the systematic creation of a toolbox of ligand-gated ion channels (LGICs) with orthogonal pharmacologic selectivity and divergent functional properties. The LGICs and their small-molecule effectors were able to activate a range of ionic conductances in genetically specified cell types. LGICs constructed for neuronal perturbation could be used to selectively manipulate neuron activity in mammalian brains in vivo. The diversity of ion channel tools accessible from this approach will be useful for examining the relationship between neuronal activity and animal behavior, as well as for cell biological and physiological applications requiring chemical control of ion conductance.
Synaptic plasticity in response to changes in physiologic state is coordinated by hormonal signals across multiple neuronal cell types, but the significance and underlying mechanisms are unclear. Here, we combine cell type-specific electrophysiological, pharmacological, and optogenetic techniques to dissect neural circuits and molecular pathways controlling synaptic plasticity onto AGRP neurons, a population that regulates feeding. We find that food deprivation elevates excitatory synaptic input, which is mediated by a presynaptic positive feedback loop involving AMP-activated protein kinase. Potentiation of glutamate release was triggered by the orexigenic hormone ghrelin and exhibited hysteresis, persisting for hours after ghrelin removal. Persistent activity was reversed by the anorexigenic hormone leptin, and optogenetic photostimulation demonstrated involvement of opioid release from POMC neurons. Based on these experiments, we propose a memory storage device for physiological state constructed from bistable synapses that are flipped between two sustained activity states by transient exposure to hormones signaling energy levels. Supported by: Howard Hughes Medical Institute.
Two intermingled hypothalamic neuron populations specified by expression of agouti-related peptide (AGRP) or pro-opiomelanocortin (POMC) positively and negatively influence feeding behavior, respectively, possibly by reciprocally regulating downstream melanocortin receptors. However, the sufficiency of these neurons to control behavior and the relationship of their activity to the magnitude and dynamics of feeding are unknown. To measure this, we used channelrhodopsin-2 for cell type-specific photostimulation. Activation of only 800 AGRP neurons in mice evoked voracious feeding within minutes. The behavioral response increased with photoexcitable neuron number, photostimulation frequency and stimulus duration. Conversely, POMC neuron stimulation reduced food intake and body weight, which required melanocortin receptor signaling. However, AGRP neuron-mediated feeding was not dependent on suppressing this melanocortin pathway, indicating that AGRP neurons directly engage feeding circuits. Furthermore, feeding was evoked selectively over drinking without training or prior photostimulus exposure, which suggests that AGRP neurons serve a dedicated role coordinating this complex behavior.
Understanding the structure and function of neural circuits are central questions in neuroscience research. To address these questions, new genetically encoded tools have been developed for mapping, monitoring, and manipulating neurons. Essential to implementation of these tools is their selective delivery to defined neuronal populations in the brain. This has been facilitated by recent improvements in cell type-specific transgene expression using recombinant adeno-associated viral vectors. Here, we highlight these developments and discuss areas for improvement that could further expand capabilities for neural circuit analysis.
Digital reconstruction of 3D neuron structures is an important step toward reverse engineering the wiring and functions of a brain. However, despite a number of existing studies, this task is still challenging, especially when a 3D microscopic image has low single-to-noise ratio and discontinued segments of neurite patterns.
The central actions of leptin are essential for homeostatic control of adipose tissue mass, glucose metabolism, and many autonomic and neuroendocrine systems. In the brain, leptin acts on numerous different cell types via the long-form leptin receptor (LepRb) to elicit its effects. The precise identification of leptin’s cellular targets is fundamental to understanding the mechanism of its pleiotropic central actions. We have systematically characterized LepRb distribution in the mouse brain using in situ hybridization in wildtype mice as well as by EYFP immunoreactivity in a novel LepRb-IRES-Cre EYFP reporter mouse line showing high levels of LepRb mRNA/EYFP coexpression. We found substantial LepRb mRNA and EYFP expression in hypothalamic and extrahypothalamic sites described before, including the dorsomedial nucleus of the hypothalamus, ventral premammillary nucleus, ventral tegmental area, parabrachial nucleus, and the dorsal vagal complex. Expression in insular cortex, lateral septal nucleus, medial preoptic area, rostral linear nucleus, and in the Edinger-Westphal nucleus was also observed and had been previously unreported. The LepRb-IRES-Cre reporter line was used to chemically characterize a population of leptin receptor-expressing neurons in the midbrain. Tyrosine hydroxylase and Cre reporter were found to be coexpressed in the ventral tegmental area and in other midbrain dopaminergic neurons. Lastly, the LepRb-IRES-Cre reporter line was used to map the extent of peripheral leptin sensing by central nervous system (CNS) LepRb neurons. Thus, we provide data supporting the use of the LepRb-IRES-Cre line for the assessment of the anatomic and functional characteristics of neurons expressing leptin receptor.
Understanding cortical circuits will require mapping the connections between specific populations of neurons, as well as determining the dendritic locations where the synapses occur. The dendrites of individual cortical neurons overlap with numerous types of local and long-range excitatory axons, but axodendritic overlap is not always a good predictor of actual connection strength. Here we developed an efficient channelrhodopsin-2 (ChR2)-assisted method to map the spatial distribution of synaptic inputs, defined by presynaptic ChR2 expression, within the dendritic arborizations of recorded neurons. We expressed ChR2 in two thalamic nuclei, the whisker motor cortex and local excitatory neurons and mapped their synapses with pyramidal neurons in layers 3, 5A and 5B (L3, L5A and L5B) in the mouse barrel cortex. Within the dendritic arborizations of L3 cells, individual inputs impinged onto distinct single domains. These domains were arrayed in an orderly, monotonic pattern along the apical axis: axons from more central origins targeted progressively higher regions of the apical dendrites. In L5 arborizations, different inputs targeted separate basal and apical domains. Input to L3 and L5 dendrites in L1 was related to whisker movement and position, suggesting that these signals have a role in controlling the gain of their target neurons. Our experiments reveal high specificity in the subcellular organization of excitatory circuits.
In the hypothalamic arcuate nucleus (ARC), pro-opiomelanocortin (POMC) neurons inhibit feeding and neuropeptide-Y (NPY) neurons stimulate feeding. We tested whether neurons in the ventromedial hypothalamic nucleus (VMH), a known satiety center, activate anorexigenic neuronal pathways in the ARC by projecting either excitatory synaptic inputs to POMC neurons and/or inhibitory inputs to NPY neurons. Using laser scanning photostimulation in brain slices from transgenic mice, we found that POMC and NPY neurons, which are interspersed in the ARC, are nevertheless regulated by anatomically distinct synaptic inputs. POMC neurons received strong excitatory input from the medial VMH (mVMH), whereas NPY neurons did not and, instead, received weak inhibitory input only from within the ARC. The strength of the excitatory input from the mVMH to POMC neurons was diminished by fasting. These data identify a new molecularly defined circuit that is dynamically regulated by nutritional state in a manner consistent with the known role of the VMH as a satiety center.
Modular synthesis and substrate stereocontrol were combined to furnish 18,000 diverse 1,3-dioxanes whose distribution in chemical space rivals that of a reference set of over 2,000 bioactive small molecules. Library quality was assessed at key synthetic stages, culminating in a detailed postsynthesis analysis of purity, yield, and structural characterizability, and the resynthesis of library subsets that did not meet quality standards. The importance of this analysis-resynthesis process is highlighted by the discovery of new biological probes through organismal and protein binding assays, and by determination of the building block and stereochemical basis for their bioactivity. This evaluation of a portion of the 1,3-dioxane library suggests that many additional probes for chemical genetics will be identified as the entire library becomes biologically annotated.