Filter
Associated Lab
- Aso Lab (30) Apply Aso Lab filter
- Betzig Lab (1) Apply Betzig Lab filter
- Bock Lab (2) Apply Bock Lab filter
- Branson Lab (8) Apply Branson Lab filter
- Card Lab (5) Apply Card Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Dickson Lab (2) Apply Dickson Lab filter
- Druckmann Lab (1) Apply Druckmann Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Funke Lab (1) Apply Funke Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (2) Apply Heberlein Lab filter
- Hermundstad Lab (2) Apply Hermundstad Lab filter
- Hess Lab (5) Apply Hess Lab filter
- Jayaraman Lab (6) Apply Jayaraman Lab filter
- Lippincott-Schwartz Lab (1) Apply Lippincott-Schwartz Lab filter
- Looger Lab (2) Apply Looger Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Reiser Lab (15) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Romani Lab (1) Apply Romani Lab filter
- Remove Rubin Lab filter Rubin Lab
- Saalfeld Lab (4) Apply Saalfeld Lab filter
- Scheffer Lab (7) Apply Scheffer Lab filter
- Schreiter Lab (1) Apply Schreiter Lab filter
- Simpson Lab (3) Apply Simpson Lab filter
- Singer Lab (1) Apply Singer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Stern Lab (1) Apply Stern Lab filter
- Svoboda Lab (3) Apply Svoboda Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Truman Lab (4) Apply Truman Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
- Turner Lab (5) Apply Turner Lab filter
- Zuker Lab (1) Apply Zuker Lab filter
Associated Project Team
- CellMap (1) Apply CellMap filter
- Fly Functional Connectome (4) Apply Fly Functional Connectome filter
- Fly Olympiad (3) Apply Fly Olympiad filter
- FlyEM (11) Apply FlyEM filter
- FlyLight (20) Apply FlyLight filter
- GENIE (1) Apply GENIE filter
- Transcription Imaging (1) Apply Transcription Imaging filter
Publication Date
- 2025 (4) Apply 2025 filter
- 2024 (4) Apply 2024 filter
- 2023 (5) Apply 2023 filter
- 2022 (1) Apply 2022 filter
- 2021 (4) Apply 2021 filter
- 2020 (9) Apply 2020 filter
- 2019 (6) Apply 2019 filter
- 2018 (7) Apply 2018 filter
- 2017 (15) Apply 2017 filter
- 2016 (3) Apply 2016 filter
- 2015 (16) Apply 2015 filter
- 2014 (9) Apply 2014 filter
- 2013 (5) Apply 2013 filter
- 2012 (8) Apply 2012 filter
- 2011 (4) Apply 2011 filter
- 2010 (4) Apply 2010 filter
- 2009 (2) Apply 2009 filter
- 2008 (4) Apply 2008 filter
- 2007 (2) Apply 2007 filter
- 2006 (1) Apply 2006 filter
- 2002 (1) Apply 2002 filter
- 2000 (2) Apply 2000 filter
- 1999 (1) Apply 1999 filter
- 1997 (1) Apply 1997 filter
- 1995 (2) Apply 1995 filter
- 1994 (2) Apply 1994 filter
- 1993 (2) Apply 1993 filter
- 1992 (1) Apply 1992 filter
- 1991 (2) Apply 1991 filter
- 1990 (3) Apply 1990 filter
- 1989 (2) Apply 1989 filter
- 1987 (2) Apply 1987 filter
- 1986 (1) Apply 1986 filter
- 1985 (1) Apply 1985 filter
- 1984 (1) Apply 1984 filter
- 1983 (1) Apply 1983 filter
- 1982 (2) Apply 1982 filter
- 1981 (1) Apply 1981 filter
- 1979 (1) Apply 1979 filter
- 1973 (1) Apply 1973 filter
Type of Publication
143 Publications
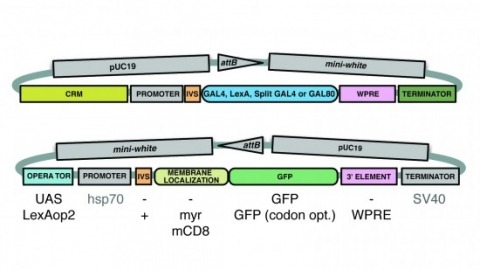
Showing 101-110 of 143 resultsA wide variety of biological experiments rely on the ability to express an exogenous gene in a transgenic animal at a defined level and in a spatially and temporally controlled pattern. We describe major improvements of the methods available for achieving this objective in Drosophila melanogaster. We have systematically varied core promoters, UTRs, operator sequences, and transcriptional activating domains used to direct gene expression with the GAL4, LexA, and Split GAL4 transcription factors and the GAL80 transcriptional repressor. The use of site-specific integration allowed us to make quantitative comparisons between different constructs inserted at the same genomic location. We also characterized a set of PhiC31 integration sites for their ability to support transgene expression of both drivers and responders in the nervous system. The increased strength and reliability of these optimized reagents overcome many of the previous limitations of these methods and will facilitate genetic manipulations of greater complexity and sophistication.
Drosophila melanogaster flies cross surmountable gaps in their walkway of widths exceeding their body length with an astounding maneuver but avoid attempts at insurmountable gaps by visual width estimation. Different mutant lines affect specific aspects of this maneuver, indicating a high complexity and modularity of the underlying motor control. Here we report on two mutants, ocelliless(1) and tay bridge(1), that, although making a correct decision to climb, fail dramatically in aiming at the right direction. Both mutants show structural defects in the protocerebral bridge, a central complex neuropil formed like a handlebar spanning the brain hemispheres. The bridge has been implicated in step-length control in walking flies and celestial E-vector orientation in locusts. In rescue experiments using tay bridge(1) flies, the integrity of the bridge was reestablished, concomitantly leading to a significant improvement of their orientation at the gap. Although producing directional scatter, their attempts were clearly aimed at the landing site. However, this partial rescue was lost in these flies at a reduced-visibility landing site. We therefore conclude that the protocerebral bridge is an essential part of a visual targeting network that transmits directional clues to the motor output via a known projection system.
To compare appetitive and aversive visual memories of the fruit fly Drosophila melanogaster, we developed a new paradigm for classical conditioning. Adult flies are trained en masse to differentially associate one of two visual conditioned stimuli (CS) (blue and green light as CS) with an appetitive or aversive chemical substance (unconditioned stimulus or US). In a test phase, flies are given a choice between the paired and the unpaired visual stimuli. Associative memory is measured based on altered visual preference in the test. If a group of flies has, for example, received a sugar reward with green light in the training, they show a significantly higher preference for the green stimulus during the test than another group of flies having received the same reward with blue light. We demonstrate critical parameters for the formation of visual appetitive memory, such as training repetition, order of reinforcement, starvation, and individual conditioning. Furthermore, we show that formic acid can act as an aversive chemical reinforcer, yielding weak, yet significant, aversive memory. These results provide a basis for future investigations into the cellular and molecular mechanisms underlying visual memory and perception in Drosophila.
Conditional expression of hairpin constructs in Drosophila is a powerful method to disrupt the activity of single genes with a spatial and temporal resolution that is impossible, or exceedingly difficult, using classical genetic methods. We previously described a method (Ni et al. 2008) whereby RNAi constructs are targeted into the genome by the phiC31-mediated integration approach using Vermilion-AttB-Loxp-Intron-UAS-MCS (VALIUM), a vector that contains vermilion as a selectable marker, an attB sequence to allow for phiC31-targeted integration at genomic attP landing sites, two pentamers of UAS, the hsp70 core promoter, a multiple cloning site, and two introns. As the level of gene activity knockdown associated with transgenic RNAi depends on the level of expression of the hairpin constructs, we generated a number of derivatives of our initial vector, called the "VALIUM" series, to improve the efficiency of the method. Here, we report the results from the systematic analysis of these derivatives and characterize VALIUM10 as the most optimal vector of this series. A critical feature of VALIUM10 is the presence of gypsy insulator sequences that boost dramatically the level of knockdown. We document the efficacy of VALIUM as a vector to analyze the phenotype of genes expressed in the nervous system and have generated a library of 2282 constructs targeting 2043 genes that will be particularly useful for studies of the nervous system as they target, in particular, transcription factors, ion channels, and transporters.
The accumulation of amyloid-beta (Abeta) into plaques is a hallmark feature of Alzheimer’s disease (AD). While amyloid precursor protein (APP)-related proteins are found in most organisms, only Abeta fragments from human APP have been shown to induce amyloid deposits and progressive neurodegeneration. Therefore, it was suggested that neurotoxic effects are a specific property of human Abeta. Here we show that Abeta fragments derived from the Drosophila orthologue APPL aggregate into intracellular fibrils, amyloid deposits, and cause age-dependent behavioral deficits and neurodegeneration. We also show that APPL can be cleaved by a novel fly beta-secretase-like enzyme. This suggests that Abeta-induced neurotoxicity is a conserved function of APP proteins whereby the lack of conservation in the primary sequence indicates that secondary structural aspects determine their pathogenesis. In addition, we found that the behavioral phenotypes precede extracellular amyloid deposit formation, supporting results that intracellular Abeta plays a key role in AD.
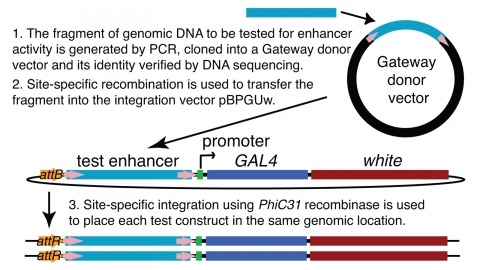
We demonstrate the feasibility of generating thousands of transgenic Drosophila melanogaster lines in which the expression of an exogenous gene is reproducibly directed to distinct small subsets of cells in the adult brain. We expect the expression patterns produced by the collection of 5,000 lines that we are currently generating to encompass all neurons in the brain in a variety of intersecting patterns. Overlapping 3-kb DNA fragments from the flanking noncoding and intronic regions of genes thought to have patterned expression in the adult brain were inserted into a defined genomic location by site-specific recombination. These fragments were then assayed for their ability to function as transcriptional enhancers in conjunction with a synthetic core promoter designed to work with a wide variety of enhancer types. An analysis of 44 fragments from four genes found that >80% drive expression patterns in the brain; the observed patterns were, on average, comprised of <100 cells. Our results suggest that the D. melanogaster genome contains >50,000 enhancers and that multiple enhancers drive distinct subsets of expression of a gene in each tissue and developmental stage. We expect that these lines will be valuable tools for neuroanatomy as well as for the elucidation of neuronal circuits and information flow in the fly brain.
Several aspects of locomotor control have been ascribed to the central complex of the insect brain; however, the role of distinct substructures of this complex is not well known. The tay bridge1 (tay1) mutant of Drosophila melanogaster was originally isolated on the basis of reduced walking speed and activity. In addition, tay1 is defective in the compensation of rotatory stimuli during walking and histologically, tay1 causes a mid-sagittal constriction of the protocerebral bridge, a constituent of the central complex. Cloning of the tay gene revealed that it encodes a novel protein with no significant homology to any known protein. To associate the behavioral phenotypes with the anatomical defect in the protocerebral bridge, we used different driver lines to express the tay cDNA in various neuronal subpopulations of the central brain in tay1-mutant flies. These experiments showed an association of the aberrant walking speed and activity with the structural defect in the protocerebral bridge. In contrast, the compensation of rotatory stimuli during walking was rescued without a restoration of the protocerebral bridge. The results of our differential rescue approach are supported by neuronal silencing experiments using conditional tetanus toxin expression in the same subset of neurons. These findings show for the first time that the walking speed and activity is controlled by different substructures of the central brain than the compensatory locomotion for rotatory stimuli.
Flexible goal-driven orientation requires that the position of a target be stored, especially in case the target moves out of sight. The capability to retain, recall and integrate such positional information into guiding behaviour has been summarized under the term spatial working memory. This kind of memory contains specific details of the presence that are not necessarily part of a long-term memory. Neurophysiological studies in primates indicate that sustained activity of neurons encodes the sensory information even though the object is no longer present. Furthermore they suggest that dopamine transmits the respective input to the prefrontal cortex, and simultaneous suppression by GABA spatially restricts this neuronal activity. Here we show that Drosophila melanogaster possesses a similar spatial memory during locomotion. Using a new detour setup, we show that flies can remember the position of an object for several seconds after it has been removed from their environment. In this setup, flies are temporarily lured away from the direction towards their hidden target, yet they are thereafter able to aim for their former target. Furthermore, we find that the GABAergic (stainable with antibodies against GABA) ring neurons of the ellipsoid body in the central brain are necessary and their plasticity is sufficient for a functional spatial orientation memory in flies. We also find that the protein kinase S6KII (ignorant) is required in a distinct subset of ring neurons to display this memory. Conditional expression of S6KII in these neurons only in adults can restore the loss of the orientation memory of the ignorant mutant. The S6KII signalling pathway therefore seems to be acutely required in the ring neurons for spatial orientation memory in flies.
The conditional expression of hairpin constructs in Drosophila melanogaster has emerged in recent years as a method of choice in functional genomic studies. To date, upstream activating site-driven RNA interference constructs have been inserted into the genome randomly using P-element-mediated transformation, which can result in false negatives due to variable expression. To avoid this problem, we have developed a transgenic RNA interference vector based on the phiC31 site-specific integration method.