Filter
Associated Lab
- Aguilera Castrejon Lab (15) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (11) Apply Ahrens Lab filter
- Baker Lab (19) Apply Baker Lab filter
- Betzig Lab (12) Apply Betzig Lab filter
- Beyene Lab (5) Apply Beyene Lab filter
- Bock Lab (3) Apply Bock Lab filter
- Branson Lab (3) Apply Branson Lab filter
- Card Lab (6) Apply Card Lab filter
- Cardona Lab (19) Apply Cardona Lab filter
- Chklovskii Lab (3) Apply Chklovskii Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Darshan Lab (4) Apply Darshan Lab filter
- Dennis Lab (1) Apply Dennis Lab filter
- Dickson Lab (14) Apply Dickson Lab filter
- Druckmann Lab (4) Apply Druckmann Lab filter
- Dudman Lab (12) Apply Dudman Lab filter
- Egnor Lab (7) Apply Egnor Lab filter
- Espinosa Medina Lab (4) Apply Espinosa Medina Lab filter
- Fetter Lab (10) Apply Fetter Lab filter
- Fitzgerald Lab (13) Apply Fitzgerald Lab filter
- Gonen Lab (32) Apply Gonen Lab filter
- Grigorieff Lab (28) Apply Grigorieff Lab filter
- Harris Lab (10) Apply Harris Lab filter
- Heberlein Lab (81) Apply Heberlein Lab filter
- Hermundstad Lab (4) Apply Hermundstad Lab filter
- Hess Lab (3) Apply Hess Lab filter
- Jayaraman Lab (4) Apply Jayaraman Lab filter
- Johnson Lab (5) Apply Johnson Lab filter
- Kainmueller Lab (19) Apply Kainmueller Lab filter
- Karpova Lab (1) Apply Karpova Lab filter
- Keleman Lab (5) Apply Keleman Lab filter
- Keller Lab (15) Apply Keller Lab filter
- Koay Lab (16) Apply Koay Lab filter
- Lavis Lab (12) Apply Lavis Lab filter
- Lee (Albert) Lab (5) Apply Lee (Albert) Lab filter
- Leonardo Lab (4) Apply Leonardo Lab filter
- Li Lab (24) Apply Li Lab filter
- Lippincott-Schwartz Lab (72) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (5) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (5) Apply Liu (Zhe) Lab filter
- Looger Lab (1) Apply Looger Lab filter
- Magee Lab (18) Apply Magee Lab filter
- Menon Lab (6) Apply Menon Lab filter
- Murphy Lab (7) Apply Murphy Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (12) Apply Otopalik Lab filter
- Pachitariu Lab (12) Apply Pachitariu Lab filter
- Pastalkova Lab (13) Apply Pastalkova Lab filter
- Pavlopoulos Lab (12) Apply Pavlopoulos Lab filter
- Pedram Lab (11) Apply Pedram Lab filter
- Reiser Lab (6) Apply Reiser Lab filter
- Riddiford Lab (24) Apply Riddiford Lab filter
- Romani Lab (12) Apply Romani Lab filter
- Rubin Lab (38) Apply Rubin Lab filter
- Saalfeld Lab (17) Apply Saalfeld Lab filter
- Satou Lab (15) Apply Satou Lab filter
- Schreiter Lab (17) Apply Schreiter Lab filter
- Sgro Lab (20) Apply Sgro Lab filter
- Simpson Lab (5) Apply Simpson Lab filter
- Singer Lab (43) Apply Singer Lab filter
- Spruston Lab (36) Apply Spruston Lab filter
- Stern Lab (83) Apply Stern Lab filter
- Sternson Lab (7) Apply Sternson Lab filter
- Stringer Lab (3) Apply Stringer Lab filter
- Svoboda Lab (4) Apply Svoboda Lab filter
- Tebo Lab (24) Apply Tebo Lab filter
- Tillberg Lab (3) Apply Tillberg Lab filter
- Tjian Lab (47) Apply Tjian Lab filter
- Truman Lab (30) Apply Truman Lab filter
- Turaga Lab (12) Apply Turaga Lab filter
- Turner Lab (11) Apply Turner Lab filter
- Wang (Shaohe) Lab (19) Apply Wang (Shaohe) Lab filter
- Wu Lab (1) Apply Wu Lab filter
- Zlatic Lab (2) Apply Zlatic Lab filter
- Zuker Lab (20) Apply Zuker Lab filter
Associated Project Team
Publication Date
- 2024 (1) Apply 2024 filter
- 2023 (1) Apply 2023 filter
- 2022 (26) Apply 2022 filter
- 2021 (19) Apply 2021 filter
- 2020 (19) Apply 2020 filter
- 2019 (25) Apply 2019 filter
- 2018 (26) Apply 2018 filter
- 2017 (31) Apply 2017 filter
- 2016 (18) Apply 2016 filter
- 2015 (57) Apply 2015 filter
- 2014 (46) Apply 2014 filter
- 2013 (58) Apply 2013 filter
- 2012 (78) Apply 2012 filter
- 2011 (92) Apply 2011 filter
- 2010 (100) Apply 2010 filter
- 2009 (102) Apply 2009 filter
- 2008 (100) Apply 2008 filter
- 2007 (85) Apply 2007 filter
- 2006 (89) Apply 2006 filter
- 2005 (67) Apply 2005 filter
- 2004 (57) Apply 2004 filter
- 2003 (58) Apply 2003 filter
- 2002 (39) Apply 2002 filter
- 2001 (28) Apply 2001 filter
- 2000 (29) Apply 2000 filter
- 1999 (14) Apply 1999 filter
- 1998 (18) Apply 1998 filter
- 1997 (16) Apply 1997 filter
- 1996 (10) Apply 1996 filter
- 1995 (18) Apply 1995 filter
- 1994 (12) Apply 1994 filter
- 1993 (10) Apply 1993 filter
- 1992 (6) Apply 1992 filter
- 1991 (11) Apply 1991 filter
- 1990 (11) Apply 1990 filter
- 1989 (6) Apply 1989 filter
- 1988 (1) Apply 1988 filter
- 1987 (7) Apply 1987 filter
- 1986 (4) Apply 1986 filter
- 1985 (5) Apply 1985 filter
- 1984 (2) Apply 1984 filter
- 1983 (2) Apply 1983 filter
- 1982 (3) Apply 1982 filter
- 1981 (3) Apply 1981 filter
- 1980 (1) Apply 1980 filter
- 1979 (1) Apply 1979 filter
- 1976 (2) Apply 1976 filter
- 1973 (1) Apply 1973 filter
- 1970 (1) Apply 1970 filter
- 1967 (1) Apply 1967 filter
Type of Publication
- Remove Non-Janelia filter Non-Janelia
1417 Publications
Showing 1361-1370 of 1417 resultsThe expression of GABA is restricted to the progeny of only six of the 24 identified postembryonic lineages in the thoracic ganglia of the tobacco hornworm, Manduca sexta (Witten and Truman, 1991). It is colocalized with a peptide similar to molluscan small cardioactive peptide B (SCPB) in some of the neurons in two of the six lineages. By combining chemical ablation of the neuroblasts at specific larval stages with birth dating of the progeny, we tested whether the expression of GABA and the SCPB-like peptide was determined strictly by cell lineage or involved cellular interactions among the members of individual clonal groups. Chemical ablation of the six specific neuroblasts that produced the GABA-positive neurons (E, K, M, N, T, and X) or of the two that produced the GABA + SCPB-like-immunoreactive neurons (K, M) prior to the generation of their lineages resulted in the loss of these immunoreactivities. These results suggest that regulation between lineages did not occur. Ablation of the K and M neuroblasts after they had produced a small portion of their lineages had no effect on the expression of GABA, but did affect the pattern of the SCPB-like immunoreactivity. Combining birth-dating techniques with transmitter immunocytochemistry revealed that it was the position in the birth order and not interactions among the clonally related neurons that influenced the peptidergic phenotype. These results suggest that cell lineage is involved in establishing the GABAergic phenotype and that both cell lineage and birth order influence the determination of the peptidergic phenotype.(ABSTRACT TRUNCATED AT 250 WORDS)
Human mitochondrial transcription factor 1 (mtTF1) has been sequenced and is a nucleus-encoded DNA binding protein of 204 amino acids (24,400 daltons). Expression of human mtTF1 in bacteria yields a protein with correct physical properties and the ability to activate mitochondrial DNA promoters. Analysis of the protein’s sequence reveals no similarities to any other DNA binding proteins except for the existence of two domains that are characteristic of high mobility group (HMG) proteins. Human mtTF1 is most closely related to a DNA binding HMG-box region in hUBF, a human protein known to be important for transcription by RNA polymerase I.
Defects in mitochondrial DNA (mtDNA) are associated with several different human diseases, including the mitochondrial encephalomyopathies. The mutations include deletions but also duplications and point mutations. Individuals with MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) carry a common A-to-G substitution in a highly conserved portion of the gene for transfer RNA(Leu(UUR)). Although the MELAS mutation may be comparable to the defect in the tRNA(Lys) gene associated with MERRF (myoclonus epilepsy associated with ragged-red fibres), it is also embedded in the middle of a tridecamer sequence necessary for the formation of the 3’ ends of 16S ribosomal RNA in vitro. We found that the MELAS mutation results in severe impairment of 16S rRNA transcription termination, which correlates with a reduced affinity of the partially purified termination protein for the MELAS template. This suggests that the molecular defect in MELAS is the inability to produce the correct type and quantity of rRNA relative to other mitochondrial gene products.
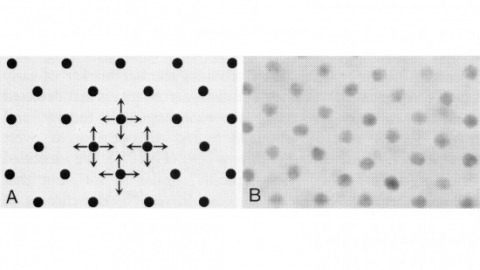
In near-field scanning optical microscopy, a light source or detector with dimensions less than the wavelength (lambda) is placed in close proximity (lambda/50) to a sample to generate images with resolution better than the diffraction limit. A near-field probe has been developed that yields a resolution of approximately 12 nm ( approximately lambda/43) and signals approximately 10(4)- to 10(6)-fold larger than those reported previously. In addition, image contrast is demonstrated to be highly polarization dependent. With these probes, near-field microscopy appears poised to fulfill its promise by combining the power of optical characterization methods with nanometric spatial resolution.
In near-field scanning optical microscopy, a light source or detector with dimensions less than the wavelength (lambda) is placed in close proximity (lambda/50) to a sample to generate images with resolution better than the diffraction limit. A near-field probe has been developed that yields a resolution of approximately 12 nm ( approximately lambda/43) and signals approximately 10(4)- to 10(6)-fold larger than those reported previously. In addition, image contrast is demonstrated to be highly polarization dependent. With these probes, near-field microscopy appears poised to fulfill its promise by combining the power of optical characterization methods with nanometric spatial resolution.
Commentary: Introduced the adiabatically tapered single mode fiber probe to near-field scanning optical microscopy which, together with shear force feedback, made the technique a practical reality. Although earlier claims of superresolution via near-field microscopy existed for nearly a decade, this paper was the first to convincingly break Abbe’s limit with visible light, as demonstrated by reproducibly resolving known, complex nanoscale patterns having features separated by much less than the wavelength. Whereas our fiber probe and shear force technologies were soon widely adopted and key to many novel applications (see above), the earlier methods proved to be technological dead ends, never achieving the results of their original claims. This experience taught me the most valuable lesson of my career: while it’s bad to bullshit others, it’s even worse to bullshit yourself. It’s a lesson sadly unheeded by many current practitioners of superresolution microscopy.
Male orchid bees of the species Eulaema meriana buzz their wings while stationary at territory perches. During buzzing, wings are first positioned laterally and then moved in a plane parallel to the ground, which probably generates a substantial airflow past the body. Within a perching episode, the ratio of buzz to pause duration decreases nonlinearly. The incidence of wing buzzing increases with ambient temperature and with duration of activity. Bees never defended territories when ambient temperatures exceeded 28.5°C. Wing buzzing may be a visual or acoustic display to conspecifics, although the brightly colored abdomen is never obscured by the wings during buzzing, and the sounds of wing buzzing are low in amplitude. The increase in buzzing frequency with increased ambient temperature and the nonlinear decrease in buzz to pause duration during perching suggest that wing buzzing may be a thermoregulatory mechanism.
In the development of multicellular organisms a diversity of cell types differentiate at specific positions. Spacing patterns, in which an array of two or more cell types forms from a uniform field of cells, are a common feature of development. Identical precursor cells may adopt different fates because of competition and inhibition between them. Such a pattern in the developing Drosophila eye is the evenly spaced array of R8 cells, around which other cell types are subsequently recruited. Genetic studies suggest that the scabrous mutation disrupts a signal produced by R8 cells that inhibits other cells from also becoming R8 cells. The scabrous locus was cloned, and it appears to encode a secreted protein partly related to the beta and gamma chains of fibrinogen. It is proposed that the sca locus encodes a lateral inhibitor of R8 differentiation. The roles of the Drosophila EGF-receptor homologue (DER) and Notch genes in this process were also investigated.
A nuclear gene (QCR9) encoding the 7.3-kDa subunit 9 of the mitochondrial cytochrome bc1 complex from Saccharomyces cerevisiae has been isolated from a yeast genomic library by hybridization with a degenerate oligonucleotide corresponding to nine amino acids proximal to the N terminus of purified subunit 9. QCR9 includes a 195-base pair open reading frame capable of encoding a protein of 66 amino acids and having a predicted molecular weight of 7471. The N-terminal methionine of subunit 9 is removed posttranslationally because the N-terminal sequence of the purified protein begins with serine 2. The ATG triplet corresponding to the N-terminal methionine is separated from the open reading frame by an intron. The intron is 213 base pairs long and contains previously reported 5’ donor, 3’ acceptor, and TACTAAC sequences necessary for splicing. The splice junctions, as well as the 5’ end of the message, were confirmed by isolation and sequencing of a cDNA copy of QCR9. In addition, the intron contains a nucleotide sequence in which 15 out of 18 nucleotides are identical with a sequence in the intron of COX4, the nuclear gene encoding cytochrome c oxidase subunit 4. The deduced amino acid sequence of the yeast subunit 9 is 39% identical with that of a protein of similar molecular weight from beef heart cytochrome bc1 complex. If conservative substitutions are allowed for, the two proteins are 56% similar. The predicted secondary structure of the 7.3-kDa protein revealed a single possible transmembrane helix, in which the amino acids conserved between beef heart and yeast are asymmetrically arranged along one face of the helix, implying that this domain of the protein is involved in a conserved interaction with another hydrophobic protein of the cytochrome bc1 complex. Two yeast strains, JDP1 and JDP2, were constructed in which QCR9 was deleted. Both strains grew very poorly, or not at all, on nonfermentable carbon sources and exhibited, at most, only 5% of wild-type ubiquinol-cytochrome c oxidoreductase activity. Optical spectra of mitochondrial membranes from the deletion strains revealed slightly reduced levels of cytochrome b. When JDP1 and JDP2 were complemented with a plasmid carrying QCR9, the resulting yeast grew normally on ethanol/glycerol and exhibited normal cytochrome c reductase activities and optical spectra. These results indicate that QCR9 encodes a 7.3-kDa subunit of the bc1 complex that is required for formation of a fully functional complex.(ABSTRACT TRUNCATED AT 400 WORDS)