Filter
Associated Lab
- Aso Lab (2) Apply Aso Lab filter
- Bock Lab (1) Apply Bock Lab filter
- Branson Lab (1) Apply Branson Lab filter
- Card Lab (3) Apply Card Lab filter
- Cardona Lab (1) Apply Cardona Lab filter
- Chklovskii Lab (1) Apply Chklovskii Lab filter
- Dickson Lab (1) Apply Dickson Lab filter
- Fetter Lab (3) Apply Fetter Lab filter
- Funke Lab (3) Apply Funke Lab filter
- Harris Lab (1) Apply Harris Lab filter
- Hess Lab (9) Apply Hess Lab filter
- Jayaraman Lab (2) Apply Jayaraman Lab filter
- Reiser Lab (2) Apply Reiser Lab filter
- Rubin Lab (11) Apply Rubin Lab filter
- Saalfeld Lab (2) Apply Saalfeld Lab filter
- Scheffer Lab (20) Apply Scheffer Lab filter
- Stern Lab (1) Apply Stern Lab filter
- Truman Lab (1) Apply Truman Lab filter
- Turner Lab (1) Apply Turner Lab filter
Associated Project Team
Publication Date
- 2025 (2) Apply 2025 filter
- 2024 (4) Apply 2024 filter
- 2023 (5) Apply 2023 filter
- 2020 (3) Apply 2020 filter
- 2019 (3) Apply 2019 filter
- 2018 (4) Apply 2018 filter
- 2017 (4) Apply 2017 filter
- 2016 (1) Apply 2016 filter
- 2015 (7) Apply 2015 filter
- 2014 (11) Apply 2014 filter
- 2013 (3) Apply 2013 filter
- 2012 (2) Apply 2012 filter
- 2010 (4) Apply 2010 filter
Type of Publication
53 Publications
Showing 41-50 of 53 resultsPixel and superpixel classifiers have become essential tools for EM segmentation algorithms. Training these classifiers remains a major bottleneck primarily due to the requirement of completely annotating the dataset which is tedious, error-prone and costly. In this paper, we propose an interactive learning scheme for the superpixel classifier for EM segmentation. Our algorithm is "active semi-supervised" because it requests the labels of a small number of examples from user and applies label propagation technique to generate these queries. Using only a small set (<20%) of all datapoints, the proposed algorithm consistently generates a classifier almost as accurate as that estimated from a complete groundtruth. We provide segmentation results on multiple datasets to show the strength of these classifiers.
This paper proposes a novel agglomerative framework for Electron Microscopy (EM) image (or volume) segmentation. For the overall segmentation methodology, we propose a context-aware algorithm that clusters the over-segmented regions of different sub-classes (representing different biological entities) in different stages. Furthermore, a delayed scheme for agglomerative clustering, which postpones the merge of newly formed bodies, is also proposed to generate a more confident boundary prediction. We report significant improvements in both segmentation accuracy and speed attained by the proposed approaches over existing standard methods on both 2D and 3D datasets.
The aim in high-resolution connectomics is to reconstruct complete neuronal connectivity in a tissue. Currently, the only technology capable of resolving the smallest neuronal processes is electron microscopy (EM). Thus, a common approach to network reconstruction is to perform (error-prone) automatic segmentation of EM images, followed by manual proofreading by experts to fix errors. We have developed an algorithm and software library to not only improve the accuracy of the initial automatic segmentation, but also point out the image coordinates where it is likely to have made errors. Our software, called gala (graph-based active learning of agglomeration), improves the state of the art in agglomerative image segmentation. It is implemented in Python and makes extensive use of the scientific Python stack (numpy, scipy, networkx, scikit-learn, scikit-image, and others). We present here the software architecture of the gala library, and discuss several designs that we consider would be generally useful for other segmentation packages. We also discuss the current limitations of the gala library and how we intend to address them.
Recent results have shown the possibility of both reconstructing connectomes of small but biologically interesting circuits and extracting from these connectomes insights into their function. However, these reconstructions were heroic proof-of-concept experiments, requiring person-months of effort per neuron reconstructed, and will not scale to larger circuits, much less the brains of entire animals. In this paper we examine what will be required to generate and use substantially larger connectomes, finding five areas that need increased attention: firstly, imaging better suited to automatic reconstruction, with excellent z-resolution; secondly, automatic detection, validation, and measurement of synapses; thirdly, reconstruction methods that keep and use uncertainty metrics for every object, from initial images, through segmentation, reconstruction, and connectome queries; fourthly, processes that are fully incremental, so that the connectome may be used before it is fully complete; and finally, better tools for analysis of connectomes, once they are obtained.
We aim to improve segmentation through the use of machine learning tools during region agglomeration. We propose an active learning approach for performing hierarchical agglomerative segmentation from superpixels. Our method combines multiple features at all scales of the agglomerative process, works for data with an arbitrary number of dimensions, and scales to very large datasets. We advocate the use of variation of information to measure segmentation accuracy, particularly in 3D electron microscopy (EM) images of neural tissue, and using this metric demonstrate an improvement over competing algorithms in EM and natural images.
Animal behaviour arises from computations in neuronal circuits, but our understanding of these computations has been frustrated by the lack of detailed synaptic connection maps, or connectomes. For example, despite intensive investigations over half a century, the neuronal implementation of local motion detection in the insect visual system remains elusive. Here we develop a semi-automated pipeline using electron microscopy to reconstruct a connectome, containing 379 neurons and 8,637 chemical synaptic contacts, within the Drosophila optic medulla. By matching reconstructed neurons to examples from light microscopy, we assigned neurons to cell types and assembled a connectome of the repeating module of the medulla. Within this module, we identified cell types constituting a motion detection circuit, and showed that the connections onto individual motion-sensitive neurons in this circuit were consistent with their direction selectivity. Our results identify cellular targets for future functional investigations, and demonstrate that connectomes can provide key insights into neuronal computations.
The most established method of reconstructing neural circuits from animals involves slicing tissue very thin, then taking mosaics of electron microscope (EM) images. To trace neurons across different images and through different sections, these images must be accurately aligned, both with the others in the same section and to the sections above and below. Unfortunately, sectioning and imaging are not ideal processes - some of the problems that make alignment difficult include lens distortion, tissue shrinkage during imaging, tears and folds in the sectioned tissue, and dust and other artifacts. In addition the data sets are large (hundreds of thousands of images) and each image must be aligned with many neighbors, so the process must be automated and reliable. This paper discusses methods of dealing with these problems, with numeric results describing the accuracy of the resulting alignments.
The ability to automatically segment an image into distinct regions is a critical aspect in many visual processing applications. Because inaccuracies often exist in automatic segmentation, manual segmentation is necessary in some application domains to correct mistakes, such as required in the reconstruction of neuronal processes from microscopic images. The goal of the automated segmentation tool is traditionally to produce the highest-quality segmentation, where quality is measured by the similarity to actual ground truth, so as to minimize the volume of manual correction necessary. Manual correction is generally orders-of-magnitude more time consuming than automated segmentation, often making handling large images intractable. Therefore, we propose a more relevant goal: minimizing the turn-around time of automated/manual segmentation while attaining a level of similarity with ground truth. It is not always necessary to inspect every aspect of an image to generate a useful segmentation. As such, we propose a strategy to guide manual segmentation to the most uncertain parts of segmentation. Our contributions include 1) a probabilistic measure that evaluates segmentation without ground truth and 2) a methodology that leverages these probabilistic measures to significantly reduce manual correction while maintaining segmentation quality.
A central problem in neuroscience is reconstructing neuronal circuits on the synapse level. Due to a wide range of scales in brain architecture such reconstruction requires imaging that is both high-resolution and high-throughput. Existing electron microscopy (EM) techniques possess required resolution in the lateral plane and either high-throughput or high depth resolution but not both. Here, we exploit recent advances in unsupervised learning and signal processing to obtain high depth-resolution EM images computationally without sacrificing throughput. First, we show that the brain tissue can be represented as a sparse linear combination of localized basis functions that are learned using high-resolution datasets. We then develop compressive sensing-inspired techniques that can reconstruct the brain tissue from very few (typically 5) tomographic views of each section. This enables tracing of neuronal processes and, hence, high throughput reconstruction of neural circuits on the level of individual synapses.
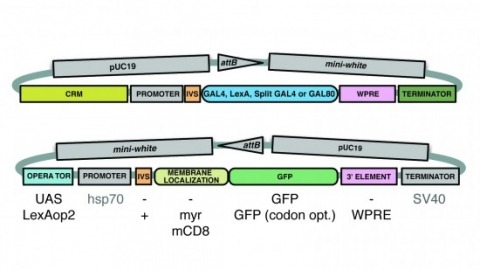
A wide variety of biological experiments rely on the ability to express an exogenous gene in a transgenic animal at a defined level and in a spatially and temporally controlled pattern. We describe major improvements of the methods available for achieving this objective in Drosophila melanogaster. We have systematically varied core promoters, UTRs, operator sequences, and transcriptional activating domains used to direct gene expression with the GAL4, LexA, and Split GAL4 transcription factors and the GAL80 transcriptional repressor. The use of site-specific integration allowed us to make quantitative comparisons between different constructs inserted at the same genomic location. We also characterized a set of PhiC31 integration sites for their ability to support transgene expression of both drivers and responders in the nervous system. The increased strength and reliability of these optimized reagents overcome many of the previous limitations of these methods and will facilitate genetic manipulations of greater complexity and sophistication.