Filter
Associated Lab
- Ahrens Lab (5) Apply Ahrens Lab filter
- Aso Lab (1) Apply Aso Lab filter
- Baker Lab (2) Apply Baker Lab filter
- Betzig Lab (4) Apply Betzig Lab filter
- Bock Lab (2) Apply Bock Lab filter
- Cardona Lab (1) Apply Cardona Lab filter
- Cui Lab (2) Apply Cui Lab filter
- Dickson Lab (3) Apply Dickson Lab filter
- Druckmann Lab (1) Apply Druckmann Lab filter
- Dudman Lab (2) Apply Dudman Lab filter
- Eddy/Rivas Lab (2) Apply Eddy/Rivas Lab filter
- Egnor Lab (1) Apply Egnor Lab filter
- Fetter Lab (3) Apply Fetter Lab filter
- Fitzgerald Lab (1) Apply Fitzgerald Lab filter
- Gonen Lab (11) Apply Gonen Lab filter
- Grigorieff Lab (4) Apply Grigorieff Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (6) Apply Heberlein Lab filter
- Hermundstad Lab (1) Apply Hermundstad Lab filter
- Hess Lab (2) Apply Hess Lab filter
- Jayaraman Lab (3) Apply Jayaraman Lab filter
- Ji Lab (1) Apply Ji Lab filter
- Johnson Lab (1) Apply Johnson Lab filter
- Karpova Lab (1) Apply Karpova Lab filter
- Keller Lab (10) Apply Keller Lab filter
- Lavis Lab (4) Apply Lavis Lab filter
- Leonardo Lab (3) Apply Leonardo Lab filter
- Lippincott-Schwartz Lab (11) Apply Lippincott-Schwartz Lab filter
- Looger Lab (10) Apply Looger Lab filter
- Magee Lab (3) Apply Magee Lab filter
- Menon Lab (3) Apply Menon Lab filter
- Pachitariu Lab (3) Apply Pachitariu Lab filter
- Pavlopoulos Lab (1) Apply Pavlopoulos Lab filter
- Reiser Lab (2) Apply Reiser Lab filter
- Riddiford Lab (5) Apply Riddiford Lab filter
- Romani Lab (1) Apply Romani Lab filter
- Rubin Lab (5) Apply Rubin Lab filter
- Satou Lab (2) Apply Satou Lab filter
- Scheffer Lab (3) Apply Scheffer Lab filter
- Schreiter Lab (7) Apply Schreiter Lab filter
- Sgro Lab (1) Apply Sgro Lab filter
- Singer Lab (9) Apply Singer Lab filter
- Spruston Lab (2) Apply Spruston Lab filter
- Stern Lab (6) Apply Stern Lab filter
- Sternson Lab (3) Apply Sternson Lab filter
- Svoboda Lab (10) Apply Svoboda Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Truman Lab (3) Apply Truman Lab filter
- Turaga Lab (2) Apply Turaga Lab filter
- Turner Lab (2) Apply Turner Lab filter
- Wu Lab (3) Apply Wu Lab filter
- Zlatic Lab (2) Apply Zlatic Lab filter
Associated Project Team
Publication Date
- December 2013 (13) Apply December 2013 filter
- November 2013 (10) Apply November 2013 filter
- October 2013 (20) Apply October 2013 filter
- September 2013 (19) Apply September 2013 filter
- August 2013 (15) Apply August 2013 filter
- July 2013 (19) Apply July 2013 filter
- June 2013 (17) Apply June 2013 filter
- May 2013 (10) Apply May 2013 filter
- April 2013 (12) Apply April 2013 filter
- March 2013 (11) Apply March 2013 filter
- February 2013 (19) Apply February 2013 filter
- January 2013 (29) Apply January 2013 filter
- Remove 2013 filter 2013
Type of Publication
194 Publications
Showing 1-10 of 194 resultsThe neural circuits that mediate behavioral choice evaluate and integrate information from the environment with internal demands and then initiate a behavioral response. Even circuits that support simple decisions remain poorly understood. In Drosophila melanogaster, oviposition on a substrate containing ethanol enhances fitness; however, little is known about the neural mechanisms mediating this important choice behavior. Here, we characterize the neural modulation of this simple choice and show that distinct subsets of dopaminergic neurons compete to either enhance or inhibit egg-laying preference for ethanol-containing food. Moreover, activity in α'β' neurons of the mushroom body and a subset of ellipsoid body ring neurons (R2) is required for this choice. We propose a model where competing dopaminergic systems modulate oviposition preference to adjust to changes in natural oviposition substrates.
In this paper, we provide a historical account of the contribution of a single line of research to our current understanding of the structure of cis-regulatory regions and the genetic basis for morphological evolution. We revisit the experiments that shed light on the evolution of larval cuticular patterns within the genus Drosophila and the evolution and structure of the shavenbaby gene. We describe the experiments that led to the discovery that multiple genetic changes in the cis-regulatory region of shavenbaby caused the loss of dorsal cuticular hairs (quaternary trichomes) in first instar larvae of Drosophila sechellia. We also discuss the experiments that showed that the convergent loss of quaternary trichomes in D. sechellia and Drosophila ezoana was generated by parallel genetic changes in orthologous enhancers of shavenbaby. We discuss the observation that multiple shavenbaby enhancers drive overlapping patterns of expression in the embryo and that these apparently redundant enhancers ensure robust shavenbaby expression and trichome morphogenesis under stressful conditions. All together, these data, collected over 13 years, provide a fundamental case study in the fields of gene regulation and morphological evolution, and highlight the importance of prolonged, detailed studies of single genes.
Neuronal computation involves the integration of synaptic inputs that are often distributed over expansive dendritic trees, suggesting the need for compensatory mechanisms that enable spatially disparate synapses to influence neuronal output. In hippocampal CA1 pyramidal neurons, such mechanisms have indeed been reported, which normalize either the ability of distributed synapses to drive action potential initiation in the axon or their ability to drive dendritic spiking locally. Here we report that these mechanisms can coexist, through an elegant combination of distance-dependent regulation of synapse number and synaptic expression of AMPA and NMDA receptors. Together, these complementary gradients allow individual dendrites in both the apical and basal dendritic trees of hippocampal neurons to operate as facile computational subunits capable of supporting both global integration in the soma/axon and local integration in the dendrite.
The hippocampal CA3 region is essential for pattern completion and generation of sharp-wave ripples. During these operations, coordinated activation of ensembles of CA3 pyramidal neurons produces spatiotemporally structured input patterns arriving onto dendrites of recurrently connected CA3 neurons. To understand how such input patterns are translated into specific output patterns, we characterized dendritic integration in CA3 pyramidal cells using two-photon imaging and glutamate uncaging. We found that thin dendrites of CA3 pyramidal neurons integrate synchronous synaptic input in a highly supralinear fashion. The amplification was primarily mediated by NMDA receptor activation and was present over a relatively broad range of spatiotemporal input patterns. The decay of voltage responses, temporal summation, and action potential output was regulated in a compartmentalized fashion mainly by a G-protein-activated inwardly rectifying K(+) current. Our results suggest that plastic dendritic integrative mechanisms may support ensemble behavior in pyramidal neurons of the hippocampal circuitry.
Freshly isolated, depolarized rat hepatocytes can repolarize into bile canalicular networks when plated in collagen sandwich cultures. We studied the events underlying this repolarization process, focusing on how hepatocytes restore ATP synthesis and resupply biosynthetic precursors after the stress of being isolated from liver. We found that soon after being plated in collagen sandwich cultures, hepatocytes converted their mitochondria into highly fused networks. This occurred through a combination of upregulation of mitochondrial fusion proteins and downregulation of a mitochondrial fission protein. Mitochondria also became more active for oxidative phosphorylation, leading to overall increased ATP levels within cells. We further observed that autophagy was upregulated in the repolarizing hepatocytes. Boosted autophagy levels likely served to recycle cellular precursors, supplying building blocks for repolarization. Repolarizing hepatocytes also extensively degraded lipid droplets, whose fatty acids provide precursors for ?-oxidation to fuel oxidative phosphorylation in mitochondria. Thus, through coordination of mitochondrial fusion, autophagy, and lipid droplet consumption, depolarized hepatocytes are able to boost ATP synthesis and biosynthetic precursors to efficiently repolarize in collagen sandwich cultures.
The gills of most teleost fishes are covered by plate-like structures, the secondary lamellae, that provide the bulk of the respiratory surface area. Water passing over the secondary lamellae exchanges gases with blood passing through the secondary lamellae, forming a system that has served as a classic model of counter-current exchange. In this study, a computational model of flow around the secondary lamellae is used to examine the hydrodynamic consequences of changes to the lamellar morphology. Consistent with previous studies, the interlamellar distance is found to strongly affect the hydrodynamic resistance of the gills. However, the presence of a small gap between the tips of the secondary lamellae is found to have a similarly strong effect on the hydrodynamic resistance and flow patterns within the gills. The results from this model have been generally formulated, allowing the calculation of the hydrodynamic resistance for measured morphometric parameters. These results provide a new basis for comparing theoretical predictions of the gill resistance with measured values, and provide a general model for examining the diversity gill morphologies observed in teleost fishes.
Biological tissue is often composed of cells with similar morphologies replicated throughout large volumes and many biological applications rely on the accurate identification of these cells and their locations from image data. Here we develop a generative model that captures the regularities present in images composed of repeating elements of a few different types. Formally, the model can be described as convolutional sparse block coding. For inference we use a variant of convolutional matching pursuit adapted to block-based representations. We extend the K-SVD learning algorithm to subspaces by retaining several principal vectors from the SVD decomposition instead of just one. Good models with little cross-talk between subspaces can be obtained by learning the blocks incrementally. We perform extensive experiments on simulated images and the inference algorithm consistently recovers a large proportion of the cells with a small number of false positives. We fit the convolutional model to noisy GCaMP6 two-photon images of spiking neurons and to Nissl-stained slices of cortical tissue and show that it recovers cell body locations without supervision. The flexibility of the block-based representation is reflected in the variability of the recovered cell shapes.
The final cleavage event that terminates cell division, abscission of the small, dense intercellular bridge, has been particularly challenging to resolve. Here, we describe imaging innovations that helped answer long-standing questions about the mechanism of abscission. We further explain how computational modeling of high-resolution data was employed to test hypotheses and generate additional insights. We present the model that emerges from application of these complimentary approaches. Similar experimental strategies will undoubtedly reveal exciting details about other underresolved cellular structures.
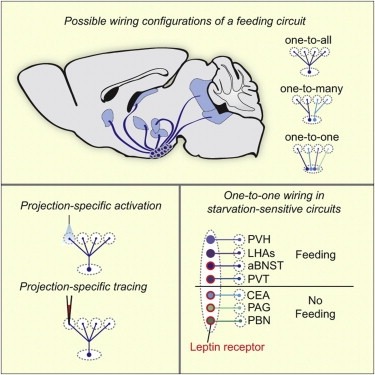
Neural circuits for essential natural behaviors are shaped by selective pressure to coordinate reliable execution of flexible goal-directed actions. However, the structural and functional organization of survival-oriented circuits is poorly understood due to exceptionally complex neuroanatomy. This is exemplified by AGRP neurons, which are a molecularly defined population that is sufficient to rapidly coordinate voracious food seeking and consumption behaviors. Here, we use cell-type-specific techniques for neural circuit manipulation and projection-specific anatomical analysis to examine the organization of this critical homeostatic circuit that regulates feeding. We show that AGRP neuronal circuits use a segregated, parallel, and redundant output configuration. AGRP neuron axon projections that target different brain regions originate from distinct subpopulations, several of which are sufficient to independently evoke feeding. The concerted anatomical and functional analysis of AGRP neuron projection populations reveals a constellation of core forebrain nodes, which are part of an extended circuit that mediates feeding behavior.
Population neural recordings with long-range temporal structure are often best understood in terms of a shared underlying low-dimensional dynamical process. Advances in recording technology provide access to an ever larger fraction of the population, but the standard computational approaches available to identify the collective dynamics scale poorly with the size of the dataset. Here we describe a new, scalable approach to discovering the low-dimensional dynamics that underlie simultaneously recorded spike trains from a neural population. Our method is based on recurrent linear models (RLMs), and relates closely to timeseries models based on recurrent neural networks. We formulate RLMs for neural data by generalising the Kalman-filter-based likelihood calculation for latent linear dynamical systems (LDS) models to incorporate a generalised-linear observation process. We show that RLMs describe motor-cortical population data better than either directly-coupled generalised-linear models or latent linear dynamical system models with generalised-linear observations. We also introduce the cascaded linear model (CLM) to capture low-dimensional instantaneous correlations in neural populations. The CLM describes the cortical recordings better than either Ising or Gaussian models and, like the RLM, can be fit exactly and quickly. The CLM can also be seen as a generalization of a low-rank Gaussian model, in this case factor analysis. The computational tractability of the RLM and CLM allow both to scale to very high-dimensional neural data.