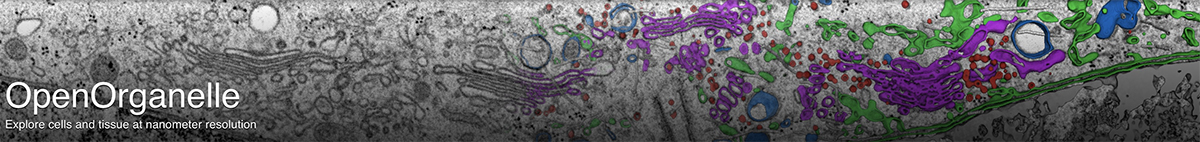
Background
 With the increasing availability of large-volume, high-resolution datasets made possible via Focused Ion Beam-Scanning Electron Microscopy (FIB-SEM) there is also an expanding need to extract the multitude of information stored within these data-rich volumes. In order to analyze these data, segmentation is first required. However, a purely manual segmentation is too time-consuming for large volumes. Thus, it is unfeasible to segment every substructure within an entire FIB-SEM volume, and an opportunity is missed.
With the increasing availability of large-volume, high-resolution datasets made possible via Focused Ion Beam-Scanning Electron Microscopy (FIB-SEM) there is also an expanding need to extract the multitude of information stored within these data-rich volumes. In order to analyze these data, segmentation is first required. However, a purely manual segmentation is too time-consuming for large volumes. Thus, it is unfeasible to segment every substructure within an entire FIB-SEM volume, and an opportunity is missed.
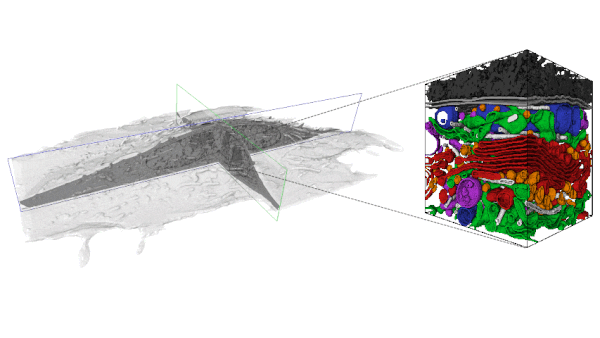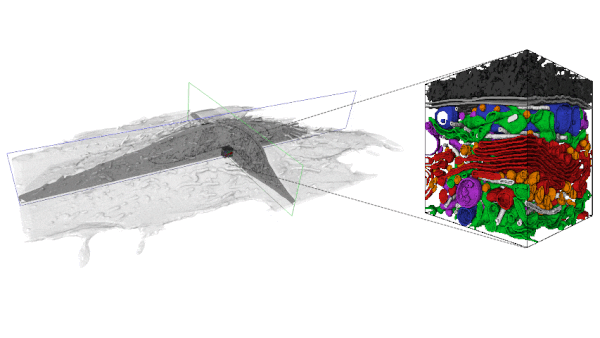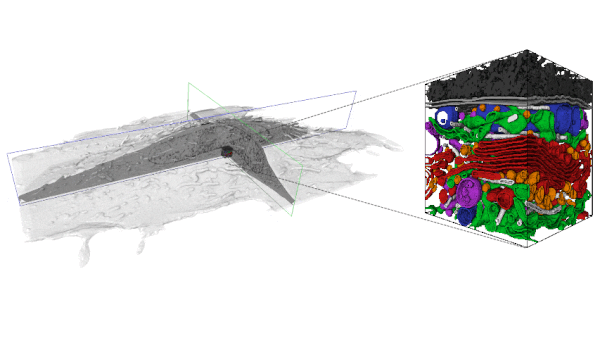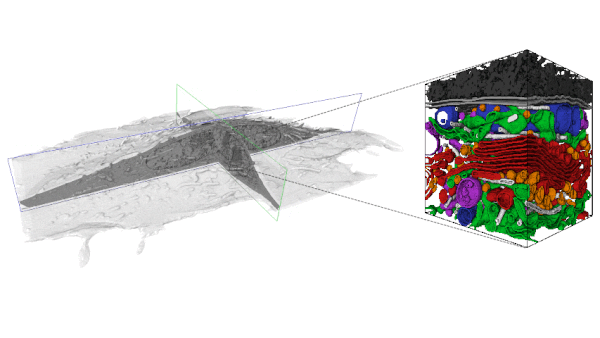
We are developing tools for automated identification of all intracellular substructures within isotropic FIB-SEM data. We trained a deep neural network to directly and simultaneously predict signed boundary distances to the nearest object boundary of 36 classes of cellular substructures. Naive thresholding of these predictions at zero produces a promising initial segmentation of cellular substructures, while different thresholds form a component tree. As a convenient side effect, boundary distance predictions also allow to immediately extract the distance between object instances, providing organelle-organelle contact sites for free. This project provides a platform for the automated segmentation and analysis of numerous FIB-SEM datasets across many cell types and species.
Pipeline
COSEM is a highly collaborative and agile team with a strategic and scalable pipeline in place.

We begin with raw EM data which is then pre-processed to remove imaging artifacts. From there skilled annotators carefully classify every voxel within a volume. These segmentations are fed into machine learning algorithms as training data. The prediction output from these algorithms are refined, proofread, and iteratively improved upon.
Once the predicted, whole-cell segmentations are achieved, quantitative analytics of subcellular distributions, interactions, sizes, and morphologies can be acquired.
Lastly, all of this data and software is publicly available for viewing and download.
We invite you to visit our data portal OpenOrganelle and Github page to explore our data and software.
OpenOrganelle
Descriptions of datasets and visualization tools can be found at openorganelle.janelia.org