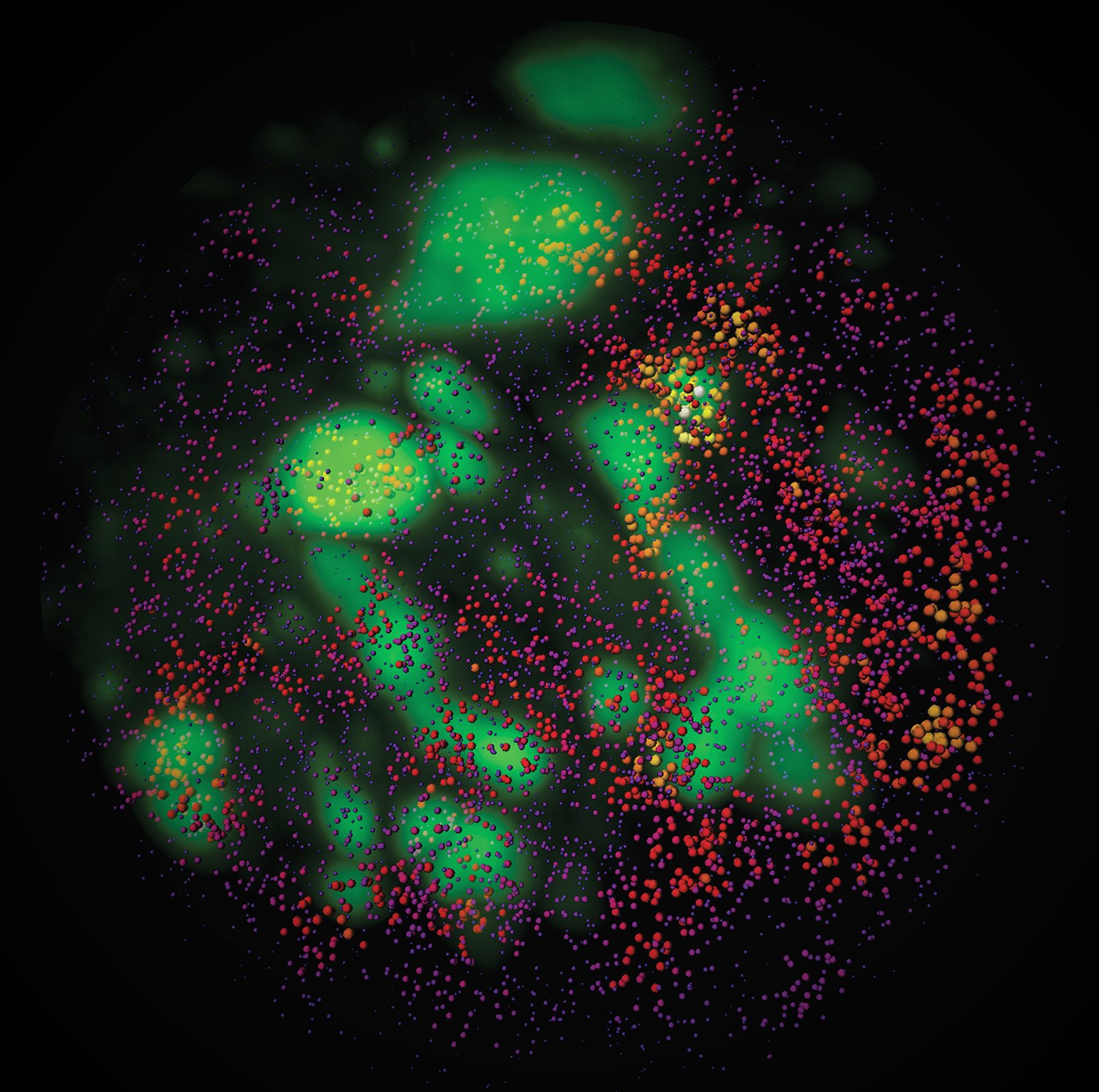
cycleHCR: spatial mapping of RNA and Proteins
Highly specific and versatile 3D imaging modality
Mapping the locations of RNA in cells and tissues is a valuable technique in biomedical research for understanding fundamental principles and developing assays. Several spatial omics techniques have demonstrated the significance of that knowledge: hybridization methods such as MERFISH and seqFISH, along with sequencing methods like FISSEQ and STARmap, have led to remarkable discoveries. Hybridization Chain Reaction (HCR) methods enhanced their results through high specificity and single-shot imaging, and they also enable deep tissue imaging.
cycleHCR overcomes the traditional limitations of fluorescence multiplexing by using multicycle DNA barcoding combined with robust HCR amplification. This allows highly multiplexed RNA and protein detection in thick tissues without the need for cross-round decoding. The technique demonstrated 254-gene transcriptome imaging in whole-mount mouse embryos and simultaneous RNA/protein mapping in hippocampal slices.

Figure 1. Representation of the cycleHCR probes and imaging
Advantages:
- Up to ~500 μm deep imaging
- Supports up to 900 barcode combinations per color channel, enabling detection of up to 2,700 unique targets across three fluorescence channels.
- Bypasses the issue of molecular crowding
- Fully automated with robotic fluidics and imaging
Applications:
- Transcriptomic profiling in whole embryos
- Subcellular protein localization with Expansion Microscopy
- RNA/protein co-mapping in brain slices
- Diagnostic mapping in pathological tissues
How it Works:
- The technique involves multiple rounds (cycles) of imaging where different molecular targets are labeled and visualized sequentially.
- cycleHCR uses stable 45-bp primary probes that remain hybridized throughout multiple imaging cycles. In each cycle, specific L+R readout probes with split HCR initiators are applied and later stripped chemically, enabling multi-round imaging without the need to reapply primary probes.
cycleHCR supports deep-tissue imaging up to 500 µm with minimal signal loss, and the platform integrates automated fluidics, robotic barcode handling, and scalable image analysis workflows.
Reference: Valentina Gandin et al., Deep-tissue transcriptomics and subcellular imaging at high spatial resolution. Science 388, eadq2084(2025). http://doi.org/10.1126/science.adq2084
IP Coverage: US Provisional Patent Application 63/643,155
Opportunity: The technology is available for internal and commercial license.
The analysis codes are freely available at GitHub and cycleHCR barcode probes are available in the paper: https://pmc.ncbi.nlm.nih.gov/articles/PMC12005972/
Tech ID: 2024-012