GAL4 Fly Lines
About the Innovation
In January 2011, the Rubin lab began depositing our collection of GAL4 driver lines in the Bloomington Stock Center at the rate of approximately 100 lines a week, and all lines were available for distribution by December 31, 2012. Data relating to these lines is available through the article and weblink (above and following):
http://www.sciencedirect.com/science/article/pii/S2211124712002926
Abstract:
We demonstrate the feasibility of generating thousands of transgenic Drosophila melanogaster lines in which the expression of an exogenous gene is reproducibly directed to distinct small subsets of cells in the adult brain. We expect the expression patterns produced by the collection of 5,000 lines that we are currently generating to encompass all neurons in the brain in various intersecting patterns.
Overlapping 3-kb DNA fragments from the flanking noncoding and intronic regions of genes thought to have patterned expression in the adult brain were inserted into a defined genomic location by site-specific recombination. These fragments were then assayed for their ability to function as transcriptional enhancers in conjunction with a synthetic core promoter designed to work with a wide variety of enhancer types. An analysis of 44 fragments from four genes found that >80% drive expression patterns in the brain; the observed patterns were, on average, comprised of <100 cells.
Our results suggest that the D. melanogaster genome contains >50,000 enhancers and that multiple enhancers drive distinct subsets of expression of a gene in each tissue and developmental stage. We expect that these lines will be valuable tools for neuroanatomy and the elucidation of neuronal circuits and information flow in the fly brain.
The methods and results described in this section were first reported in Pfeiffer et al. 2008 ( http://www.pnas.org/content/105/28/9715.long ), where more details can be found. Our approach is based on generating lines in which the expression of GAL4 is driven by a defined DNA fragment that contains one or more enhancers. The pipeline we are using to produce the lines is shown in the first diagram, and an example of the analysis as applied to the octopamine receptor 2 gene is shown second.
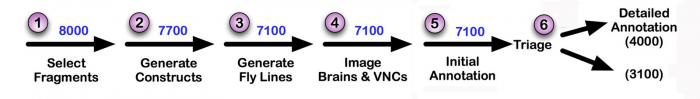
Pipeline for GAL4 driver production and imaging. (1) We selected approximately 1200 genes for which available expression data or predicted function implies expression in a subset of cells in the adult brain: for example, genes encoding transcription factors, neuropeptides, cell surface proteins, ion channels, transporters, and receptors. We spanned the flanking upstream and downstream intergenic regions of these genes, as well as any of their introns larger than 300 bp, with fragments of DNA that averaged 3 kb in length and overlapped (in regions that a single fragment could not cover) by about 1 kb. (2) We generated 7,800 fragments by PCR from genomic DNA which were then cloned, sequence verified and inserted upstream of a core promoter and the GAL4 coding region. In about 200 cases where the upstream intergenic region was small, we generated PCR fragments that also contained the start site of transcription and used them to create transcriptional fusion constructs. (3) Constructs were inserted into the attP2 integration site using the phiC31 site-specific integration system and homozygous stocks generated; Genetic Services, Inc performed the injections, and the Janelia Fly Facility generated and maintains the homozygous stocks. (4) GAL4 driver lines were crossed to a UAS-GFP reporter, adult brains and ventral nerve cords (VNCs) were imaged by confocal microscopy by the FlyLight project team. (5) See below for a description of the annotation process. (6) Lines with either no expression (about 10%) or too broad expression (about 30%) were not analyzed further, although many of these lines show highly specific patterns in other tissues. Lines are being deposited in the Bloomington Stock Center (see Tools and Reagents) at the rate of 100 lines a week. A database showing the expression patterns of the lines and our annotations will be made public in September 2012.
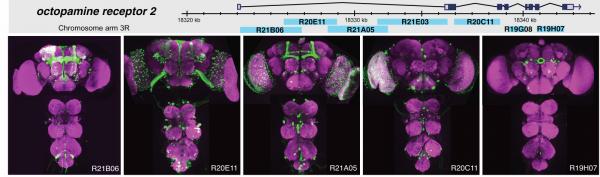
Distinct expression patterns are generated by fragments of the octopamine receptor 2 gene. (Top) Diagram of the genomic locus showing the structures of the transcription unit and the position of the fragments that were assayed for enhancer activity. (Bottom) Expression driven by the indicated five fragments in the adult brain and ventral nerve cord is shown in green, and the neuropil is counterstained in magenta.
Our preliminary analysis of the patterns observed from 7,000 lines indicates that it should be possible to establish a collection of transgenic Drosophila lines, each directing expression to a small subset of cells in the adult brain. In sum, cover all cells in the brain. About half the fragments we tested gave expression in 10 to 200 cells—0.03 to 0.7 percent of the 30,000 cells thought to comprise the central brain, a fraction of cells that should be useful for a number for anatomical, physiological or behavioral studies. We recognize that the preferred cell number for behavioral studies is unknown and will certainly depend on the particular behavior and assay; however, we expect that our lines and the experimental approaches they enable will provide the versatility needed to generate expression patterns of the desired sizes. Furthermore, we expect that the 3,000 lines we expect to generate having expression patterns within the 10 to 200 range of cell numbers will be adequate to represent the vast majority, if not all, cells in the central brain in various overlapping patterns. But they are not individually adequate to provide cell type-specific expression. To achieve that goal, we use intersectional methods as described in the section of new genetic tools for manipulating transgene expression below.
Barret Pfeiffer, Chris Murphy, and Heather Dionne led individuals to construct the lines, working with Genetic Systems, Inc. and the Fly Facility. Teri Ngo worked closely with the FlyLight histology team to generate the confocal images.
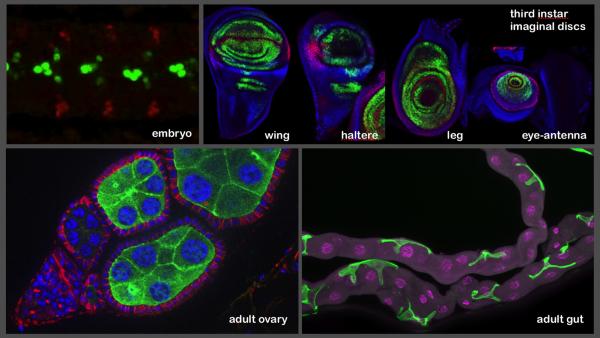
In addition to the adult brain and ventral nerve cord, the lines are being imaged in the third instar larval nervous system by the FlyLight project team, and a team led by Jim Truman analyzed the expression patterns.
As part of the Janelia visitor program, the expression patterns generated by the lines are also being imaged in the stage 17 embryonic nervous system (by the laboratory of Chris Doe), in the imaginal discs (by the laboratory of Richard Mann), and in adult ovaries and gut (by the laboratory of Allan Spradling). Images from these efforts are shown:
For inquiries, please reference:
Janelia 2008-007