Our main research interest involved deciphering the means by which the genetic information stored in DNA molecules is retrieved in a controlled and orderly fashion during transcription, the biochemical process that leads to the production of specific proteins in animal cells. We used both in vitro and in vivo systems in a multipronged approach to the problem of gene control.
For example, we devised various means of isolating the individual components of the cell responsible for transcription and of reconstructing this complex reaction in the test tube. In this way, we could study the detailed molecular mechanisms that direct the exquisitely sensitive tuning up and down of gene expression in human and animal regulatory systems. We also used cells and whole organisms (e.g., flies and mice) to interface our biochemical studies with physiologically relevant situations that require the switching on and off of specific genes during cell growth, development, and differentiation in metazoan organisms.
Developing Single-Cell Methods to Study Transcription
Although our attempts to dissect the elaborate process of transcription relied on a combination of in vitro biochemistry and in vivo reverse genetics, it was evident that a significant gap remains between in vitro mechanisms and in vivo physiology. Thus, we generated a battery of reagents and novel assays to bridge this gap. One effort was to develop assays at the single-cell level so that we could locate, track, and study transcription in individual cells. Using a combination of fluorescence microscopy, antibody staining, GFP (green fluorescent protein) tracking, ChIPs (chromatin immunoprecipitation), FISH (fluorescence in situ hybridization), and genomic arrays, we assembled tools to analyze the in vivo mechanisms that regulate transcription at the single-cell resolution. We adapted single-molecule techniques such as FRET (fluorescence resonance energy transfer) and cryo-EM (cryo-electron microscopy) to complement our functional studies of key molecular transactions during the regulation of transcription.
Proteins That Regulate Gene Expression
Among the most important families of proteins in the nucleus are the sequence-specific transcription factors that bind to DNA at select sites and regulate the expression of genes in a tissue-specific and developmentally regulated fashion. A significant technical advance was the development of biochemical methods to purify these rare and fragile proteins, which in turn allows us to isolate the genes that encode these regulatory factors.
These sequence-specific transcription factors not only regulate the temporal cascade of gene expression during development of multicellular organisms but also are often good candidates for genes that lead to cancer. For example, many of these transcription factors turn out to be either oncogenes (Jun/Fos) or tumor-suppressor genes (p53, Rb). At the same time, a growing body of evidence links specific transcription factors (such as FOXO, PPARγ, NFκB) to various metabolic diseases and inflammation, while Sp1 has been implicated in Huntington's disease.
Triggering of Transcription by Promoter-Specific Regulators
A fundamental question that remains poorly understood concerns how sequence-specific DNA-binding proteins direct transcription. To address this critical issue, our lab fractionated and isolated the multiple components necessary to reconstitute activator-responsive transcription in the test tube. In the process of dissecting the general transcriptional apparatus, we discovered previously undetected components that serve as the functional bridge between upstream regulatory proteins, such as the human transcription factor Sp1, and the initiation complex that contains RNA polymerase. These novel factors, which we have called coactivators, appear to be part of the missing link that directs promoter-selective transcription in animal cells and are likely to represent a diverse and essential class of regulatory proteins.
The first class of coactivators we identified consists of the TATA-binding protein (TBP)-associated factors (TAFIIs), which make up the core transcription complex called TFIID. Biochemical characterization of these factors revealed that transcriptional activators can bind selective TAFII subunits of the TFIID complex. The specificity and function of individual TAFIIs mediating transcriptional activation remain to be determined, however, and we continue to work toward this goal. To this end, we have used both x-ray crystallography and EM analysis to determine the three-dimensional structure of transcription proteins and large multicomponent complexes. These structure/function studies have revealed that TAFs can bind core promoter DNA, interact with selective activators, catalyze various enzymatic reactions (e.g., kinase, acetyltransferase), and target binding of TFIID to specifically acetylated nucleosomal templates.
Structural Studies of Multisubunit Transcriptional Cofactors
Our biochemical studies also identified two additional human cofactor complexes, CRSP (cofactor required for Sp1) and ARC (activator-recruited cofactor), that together with the TAFIIs help mediate transcriptional activation. Structural determination by EM and biochemical assays revealed that ARC-L and CRSP are substantially different in size and shape and display differential coactivator functions. Moreover, EM analysis of independently derived CRSP complexes reveals distinct conformations induced by different activators. These results suggest that CRSP may potentiate transcription via specific activator-induced conformational changes.
Identification of TBP-Related Factors and Cell-Type-Specific TAFIIs
Eukaryotic cells were originally thought to contain a single TBP that directs transcription by all cellular RNA polymerases. However, we recently identified two TBP-related factors, TRF-1 and TRF-2, that interact with the basal transcription factors TFIIA and TFIIB to initiate gene-selective RNA synthesis. In addition to multiple TRFs, we have also discovered tissue- or cell-type-specific TAFIIs. In vitro biochemical and in vivo genetic analyses reveal that TAFII105 is selectively expressed in the granulosa cells of the mouse ovary. Deletion of this gene by homologous recombination results in infertile female mice with defects in ovarian development due to down-regulated genes. The discovery of multiple TFIID-like complexes confirms that the core molecular machinery in metazoans has diversified to accommodate complex patterns of transcription.
Transcriptional Control and Disease
Our studies of activators and coactivators also provided insights into the molecular mechanisms underlying various human diseases. For example, recent studies suggest that during the onset of Huntington's disease, the mutant Huntingtin protein disrupts an important interaction between the human transcription factor Sp1 and its target coactivator TAFII130. Another example is the finding that type II diabetes can be controlled by a class of insulin sensitizers that modulate the specific interaction between specific coactivators and the ligand-binding domain of the PPARγ nuclear receptor. Finally, transcription factor FOXO was found to regulate the insulin receptor in a feedback mechanism controlled by insulin and nutrient deprivation. This finding provides a potential link between FOXO and insulin-resistant diabetes. As we learn more about the molecular mechanisms that have evolved to regulate transcription and disease mechanisms, opportunities for new therapeutic strategies will emerge.
Transcriptional Mechanisms Regulating Stem Cell Self-Renewal and Differentiation
A new area of research we initiated targets the identification of cell-type-specific transcription factors responsible for directing human stem cell maintenance as well as changes to the core promoter recognition apparatus during differentiation. In particular, we isolated a novel stem cell–specific cofactor complex (SCC) that is required to mediate the transcriptional activation of the NANOG gene by OCT4/SOX2 in human embryonic stem cells. In addition, our studies of skeletal muscle differentiation revealed dramatic changes in the requirement for core promoter-binding complexes necessary to drive differentiation of myoblasts into myotubes. We also studied the role of key transcription activators that drive embryonic stem cells to become dopamine-producing neurons.
A grant from the National Institutes of Health provided partial support for the work on Sp1, CRSP, ARC, and other human enhancer and basal transcription factors.
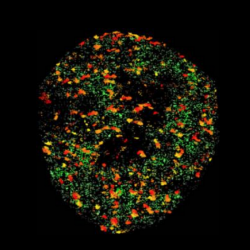 |
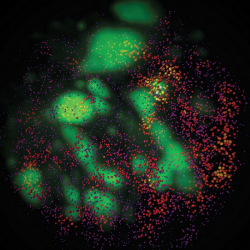 |
|
|
Pol II and Sox2 enhancer clusters in live cells. |
Sox2 enhancer clusters and Heterochromatin in live cells. |
