Filter
Associated Lab
- Aso Lab (1) Apply Aso Lab filter
- Baker Lab (3) Apply Baker Lab filter
- Betzig Lab (7) Apply Betzig Lab filter
- Bock Lab (1) Apply Bock Lab filter
- Cui Lab (3) Apply Cui Lab filter
- Dickson Lab (1) Apply Dickson Lab filter
- Dudman Lab (1) Apply Dudman Lab filter
- Eddy/Rivas Lab (5) Apply Eddy/Rivas Lab filter
- Fetter Lab (2) Apply Fetter Lab filter
- Gonen Lab (1) Apply Gonen Lab filter
- Hess Lab (2) Apply Hess Lab filter
- Jayaraman Lab (2) Apply Jayaraman Lab filter
- Ji Lab (2) Apply Ji Lab filter
- Keller Lab (2) Apply Keller Lab filter
- Lavis Lab (5) Apply Lavis Lab filter
- Lee (Albert) Lab (1) Apply Lee (Albert) Lab filter
- Leonardo Lab (2) Apply Leonardo Lab filter
- Looger Lab (7) Apply Looger Lab filter
- Magee Lab (1) Apply Magee Lab filter
- Menon Lab (3) Apply Menon Lab filter
- Murphy Lab (1) Apply Murphy Lab filter
- Reiser Lab (2) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Rubin Lab (3) Apply Rubin Lab filter
- Scheffer Lab (2) Apply Scheffer Lab filter
- Schreiter Lab (2) Apply Schreiter Lab filter
- Simpson Lab (3) Apply Simpson Lab filter
- Singer Lab (1) Apply Singer Lab filter
- Sternson Lab (6) Apply Sternson Lab filter
- Svoboda Lab (7) Apply Svoboda Lab filter
- Tjian Lab (2) Apply Tjian Lab filter
- Truman Lab (1) Apply Truman Lab filter
- Zlatic Lab (1) Apply Zlatic Lab filter
- Zuker Lab (2) Apply Zuker Lab filter
Associated Project Team
Publication Date
- December 2011 (10) Apply December 2011 filter
- November 2011 (8) Apply November 2011 filter
- October 2011 (8) Apply October 2011 filter
- September 2011 (8) Apply September 2011 filter
- August 2011 (9) Apply August 2011 filter
- July 2011 (5) Apply July 2011 filter
- June 2011 (10) Apply June 2011 filter
- May 2011 (6) Apply May 2011 filter
- April 2011 (5) Apply April 2011 filter
- March 2011 (6) Apply March 2011 filter
- February 2011 (8) Apply February 2011 filter
- January 2011 (15) Apply January 2011 filter
- Remove 2011 filter 2011
98 Janelia Publications
Showing 11-20 of 98 resultsFull reconstruction of neuron morphology is of fundamental interest for the analysis and understanding of their functioning. We have developed a novel method capable of automatically tracing neurons in three-dimensional microscopy data. In contrast to template-based methods, the proposed approach makes no assumptions about the shape or appearance of neurite structure. Instead, an efficient seeding approach is applied to capture complex neuronal structures and the tracing problem is solved by computing the optimal reconstruction with a weighted graph. The optimality is determined by the cost function designed for the path between each pair of seeds and by topological constraints defining the component interrelations and completeness. In addition, an automated neuron comparison method is introduced for performance evaluation and structure analysis. The proposed algorithm is computationally efficient and has been validated using different types of microscopy data sets including Drosophila’s projection neurons and fly neurons with presynaptic sites. In all cases, the approach yielded promising results.
High-resolution microscopic imaging of biological samples often produces multiple 3D image tiles to cover a large field of view of specimen. Usually each tile has a large size, in the range of hundreds of megabytes to several gigabytes. For many of our image data sets, existing software tools are often unable to stitch those 3D tiles into a panoramic view, thus impede further data analysis. We propose a simple, but accurate, robust, and automatic method to stitch a group of image tiles without a priori adjacency information of them. We first use a multiscale strategy to register a pair of 3D image tiles rapidly, achieving about 8~10 times faster speed and 10 times less memory requirement compared to previous methods. Then we design a minimum-spanning-tree based method to determine the optimal adjacency of tiles. We have successfully stitched large image stacks of model animals including C. elegans, fruit fly, dragonfly, and mouse, which could not be stitched by several existing methods.
Digital reconstruction of neurons from microscope images is an important and challenging problem in neuroscience. In this paper, we propose a model-based method to tackle this problem. We first formulate a model structure, then develop an algorithm for computing it by carefully taking into account morphological characteristics of neurons, as well as the image properties under typical imaging protocols. The method has been tested on the data sets used in the DIADEM competition and produced promising results for four out of the five data sets.
Motivation: Digital reconstruction, or tracing, of 3D neuron structures is critical toward reverse engineering the wiring and functions of a brain. However, despite a number of existing studies, this task is still challenging, especially when a 3D microscopic image has low signal-to-noise ratio (SNR) and fragmented neuron segments. Published work can handle these hard situations only by introducing global prior information, such as where a neurite segment starts and terminates. However, manual incorporation of such global information can be very time consuming. Thus, a completely automatic approach for these hard situations is highly desirable. Results: We have developed an automatic graph algorithm, called the all-path pruning (APP), to trace the 3D structure of a neuron. To avoid potential mis-tracing of some parts of a neuron, an APP first produces an initial over-reconstruction, by tracing the optimal geodesic shortest path from the seed location to every possible destination voxel/pixel location in the image. Since the initial reconstruction contains all the possible paths and thus could contain redundant structural components (SC), we simplify the entire reconstruction without compromising its connectedness by pruning the redundant structural elements, using a new maximal-covering minimal-redundant (MCMR) subgraph algorithm. We show that MCMR has a linear computational complexity and will converge. We examined the performance of our method using challenging 3D neuronal image datasets of model organisms (e.g. fruit fly).
Analyzing Drosophila melanogaster neural expression patterns in thousands of three-dimensional image stacks of individual brains requires registering them into a canonical framework based on a fiducial reference of neuropil morphology. Given a target brain labeled with predefined landmarks, the BrainAligner program automatically finds the corresponding landmarks in a subject brain and maps it to the coordinate system of the target brain via a deformable warp. Using a neuropil marker (the antibody nc82) as a reference of the brain morphology and a target brain that is itself a statistical average of data for 295 brains, we achieved a registration accuracy of 2 μm on average, permitting assessment of stereotypy, potential connectivity and functional mapping of the adult fruit fly brain. We used BrainAligner to generate an image pattern atlas of 2954 registered brains containing 470 different expression patterns that cover all the major compartments of the fly brain.
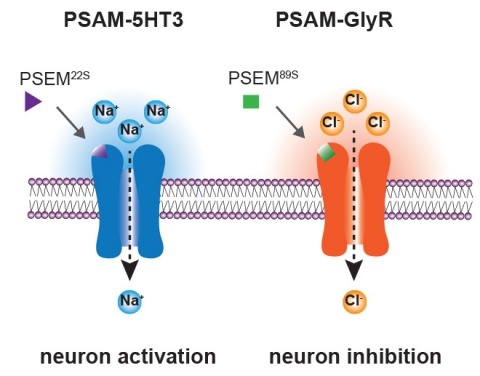
Ionic flux mediates essential physiological and behavioral functions in defined cell populations. Cell type-specific activators of diverse ionic conductances are needed for probing these effects. We combined chemistry and protein engineering to enable the systematic creation of a toolbox of ligand-gated ion channels (LGICs) with orthogonal pharmacologic selectivity and divergent functional properties. The LGICs and their small-molecule effectors were able to activate a range of ionic conductances in genetically specified cell types. LGICs constructed for neuronal perturbation could be used to selectively manipulate neuron activity in mammalian brains in vivo. The diversity of ion channel tools accessible from this approach will be useful for examining the relationship between neuronal activity and animal behavior, as well as for cell biological and physiological applications requiring chemical control of ion conductance.
Gamma-aminobutyric acid (GABA) is the major inhibitory neurotransmitter in the mammalian brain. Once released, it is removed from the extracellular space by cellular uptake catalyzed by GABA transporter proteins. Four GABA transporters (GAT1, GAT2, GAT3 and BGT1) have been identified. Inhibition of the GAT1 by the clinically available anti-epileptic drug tiagabine has been an effective strategy for the treatment of some patients with partial seizures. Recently, the investigational drug EF1502, which inhibits both GAT1 and BGT1, was found to exert an anti-convulsant action synergistic to that of tiagabine, supposedly due to inhibition of BGT1. The present study addresses the role of BGT1 in seizure control and the effect of EF1502 by developing and exploring a new mouse line lacking exons 3-5 of the BGT1 (slc6a12) gene. The deletion of this sequence abolishes the expression of BGT1 mRNA. However, homozygous BGT1-deficient mice have normal development and show seizure susceptibility indistinguishable from that in wild-type mice in a variety of seizure threshold models including: corneal kindling, the minimal clonic and minimal tonic extension seizure threshold tests, the 6Hz seizure threshold test, and the i.v. pentylenetetrazol threshold test. We confirm that BGT1 mRNA is present in the brain, but find that the levels are several hundred times lower than those of GAT1 mRNA; possibly explaining the apparent lack of phenotype. In conclusion, the present results do not support a role for BGT1 in the control of seizure susceptibility and cannot provide a mechanistic understanding of the synergism that has been previously reported with tiagabine and EF1502.
Biotinidase deficiency is the primary enzymatic defect in biotin-responsive, late-onset multiple carboxylase deficiency. Untreated children with profound biotinidase deficiency usually exhibit neurological symptoms including lethargy, hypotonia, seizures, developmental delay, sensorineural hearing loss and optic atrophy; and cutaneous symptoms including skin rash, conjunctivitis and alopecia. Although the clinical features of the disorder markedly improve or are prevented with biotin supplementation, some symptoms, once they occur, such as developmental delay, hearing loss and optic atrophy, are usually irreversible. To prevent development of symptoms, the disorder is screened for in the newborn period in essentially all states and in many countries. In order to better understand many aspects of the pathophysiology of the disorder, we have developed a transgenic biotinidase-deficient mouse. The mouse has a null mutation that results in no detectable serum biotinidase activity or cross-reacting material to antibody prepared against biotinidase. When fed a biotin-deficient diet these mice develop neurological and cutaneous symptoms, carboxylase deficiency, mild hyperammonemia, and exhibit increased urinary excretion of 3-hydroxyisovaleric acid and biotin and biotin metabolites. The clinical features are reversed with biotin supplementation. This biotinidase-deficient animal can be used to study systematically many aspects of the disorder and the role of biotinidase, biotin and biocytin in normal and in enzyme-deficient states.
Concomitant with the publication of this Special Issue of Neuroinformatics, a substantially updated version of the DIADEM web site has been released at http://diademchallenge.org. This web site was originally designed to host the challenge for automating the digital reconstruction of axonal and dendritic morphology (hence the DIADEM acronym). This post-competition version features additional content for continued use as the access point for DIADEM-related material. From the very beginning, one of the spirits of DIADEM has been to share data and resources with the neuroscience research community at large. The resources available from or linked to the DIADEM website constitute a substantial scientific legacy of the 2009/2010 competition. The new content includes finalist algorithms, image stack data, gold standard reconstructions, an updated DIADEM metric, and a retrospective on the competition in text and images.
Uncovering the direct regulatory targets of doublesex (dsx) and fruitless (fru) is crucial for an understanding of how they regulate sexual development, morphogenesis, differentiation and adult functions (including behavior) in Drosophila melanogaster. Using a modified DamID approach, we identified 650 DSX-binding regions in the genome from which we then extracted an optimal palindromic 13 bp DSX-binding sequence. This sequence is functional in vivo, and the base identity at each position is important for DSX binding in vitro. In addition, this sequence is enriched in the genomes of D. melanogaster (58 copies versus approximately the three expected from random) and in the 11 other sequenced Drosophila species, as well as in some other Dipterans. Twenty-three genes are associated with both an in vivo peak in DSX binding and an optimal DSX-binding sequence, and thus are almost certainly direct DSX targets. The association of these 23 genes with optimum DSX binding sites was used to examine the evolutionary changes occurring in DSX and its targets in insects.