Filter
Associated Lab
- Aguilera Castrejon Lab (1) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (52) Apply Ahrens Lab filter
- Aso Lab (40) Apply Aso Lab filter
- Baker Lab (19) Apply Baker Lab filter
- Betzig Lab (100) Apply Betzig Lab filter
- Beyene Lab (8) Apply Beyene Lab filter
- Bock Lab (14) Apply Bock Lab filter
- Branson Lab (49) Apply Branson Lab filter
- Card Lab (35) Apply Card Lab filter
- Cardona Lab (44) Apply Cardona Lab filter
- Chklovskii Lab (10) Apply Chklovskii Lab filter
- Clapham Lab (13) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (8) Apply Darshan Lab filter
- Dickson Lab (32) Apply Dickson Lab filter
- Druckmann Lab (21) Apply Druckmann Lab filter
- Dudman Lab (38) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (4) Apply Egnor Lab filter
- Espinosa Medina Lab (15) Apply Espinosa Medina Lab filter
- Feliciano Lab (7) Apply Feliciano Lab filter
- Fetter Lab (31) Apply Fetter Lab filter
- Fitzgerald Lab (16) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (38) Apply Funke Lab filter
- Gonen Lab (59) Apply Gonen Lab filter
- Grigorieff Lab (34) Apply Grigorieff Lab filter
- Harris Lab (50) Apply Harris Lab filter
- Heberlein Lab (13) Apply Heberlein Lab filter
- Hermundstad Lab (22) Apply Hermundstad Lab filter
- Hess Lab (73) Apply Hess Lab filter
- Ilanges Lab (2) Apply Ilanges Lab filter
- Jayaraman Lab (42) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (1) Apply Johnson Lab filter
- Karpova Lab (13) Apply Karpova Lab filter
- Keleman Lab (8) Apply Keleman Lab filter
- Keller Lab (61) Apply Keller Lab filter
- Koay Lab (2) Apply Koay Lab filter
- Lavis Lab (136) Apply Lavis Lab filter
- Lee (Albert) Lab (29) Apply Lee (Albert) Lab filter
- Leonardo Lab (19) Apply Leonardo Lab filter
- Li Lab (4) Apply Li Lab filter
- Lippincott-Schwartz Lab (95) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (1) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (56) Apply Liu (Zhe) Lab filter
- Looger Lab (137) Apply Looger Lab filter
- Magee Lab (31) Apply Magee Lab filter
- Menon Lab (12) Apply Menon Lab filter
- Murphy Lab (6) Apply Murphy Lab filter
- O'Shea Lab (5) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Pachitariu Lab (35) Apply Pachitariu Lab filter
- Pastalkova Lab (5) Apply Pastalkova Lab filter
- Pavlopoulos Lab (7) Apply Pavlopoulos Lab filter
- Pedram Lab (4) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (45) Apply Reiser Lab filter
- Riddiford Lab (20) Apply Riddiford Lab filter
- Romani Lab (31) Apply Romani Lab filter
- Rubin Lab (105) Apply Rubin Lab filter
- Saalfeld Lab (46) Apply Saalfeld Lab filter
- Satou Lab (1) Apply Satou Lab filter
- Scheffer Lab (36) Apply Scheffer Lab filter
- Schreiter Lab (50) Apply Schreiter Lab filter
- Sgro Lab (1) Apply Sgro Lab filter
- Shroff Lab (30) Apply Shroff Lab filter
- Simpson Lab (18) Apply Simpson Lab filter
- Singer Lab (37) Apply Singer Lab filter
- Spruston Lab (57) Apply Spruston Lab filter
- Stern Lab (73) Apply Stern Lab filter
- Sternson Lab (47) Apply Sternson Lab filter
- Stringer Lab (31) Apply Stringer Lab filter
- Svoboda Lab (131) Apply Svoboda Lab filter
- Tebo Lab (9) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (18) Apply Tillberg Lab filter
- Tjian Lab (17) Apply Tjian Lab filter
- Truman Lab (58) Apply Truman Lab filter
- Turaga Lab (38) Apply Turaga Lab filter
- Turner Lab (26) Apply Turner Lab filter
- Vale Lab (7) Apply Vale Lab filter
- Voigts Lab (3) Apply Voigts Lab filter
- Wang (Meng) Lab (18) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (6) Apply Wang (Shaohe) Lab filter
- Wu Lab (8) Apply Wu Lab filter
- Zlatic Lab (26) Apply Zlatic Lab filter
- Zuker Lab (5) Apply Zuker Lab filter
Associated Project Team
- CellMap (12) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- FIB-SEM Technology (2) Apply FIB-SEM Technology filter
- Fly Descending Interneuron (10) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (53) Apply FlyEM filter
- FlyLight (49) Apply FlyLight filter
- GENIE (45) Apply GENIE filter
- Integrative Imaging (3) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (18) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (26) Apply Tool Translation Team (T3) filter
- Transcription Imaging (45) Apply Transcription Imaging filter
Associated Support Team
- Project Pipeline Support (3) Apply Project Pipeline Support filter
- Anatomy and Histology (18) Apply Anatomy and Histology filter
- Cryo-Electron Microscopy (34) Apply Cryo-Electron Microscopy filter
- Electron Microscopy (15) Apply Electron Microscopy filter
- Gene Targeting and Transgenics (11) Apply Gene Targeting and Transgenics filter
- Integrative Imaging (17) Apply Integrative Imaging filter
- Invertebrate Shared Resource (40) Apply Invertebrate Shared Resource filter
- Janelia Experimental Technology (37) Apply Janelia Experimental Technology filter
- Management Team (1) Apply Management Team filter
- Molecular Genomics (15) Apply Molecular Genomics filter
- Primary & iPS Cell Culture (14) Apply Primary & iPS Cell Culture filter
- Project Technical Resources (47) Apply Project Technical Resources filter
- Quantitative Genomics (19) Apply Quantitative Genomics filter
- Scientific Computing Software (91) Apply Scientific Computing Software filter
- Scientific Computing Systems (6) Apply Scientific Computing Systems filter
- Viral Tools (14) Apply Viral Tools filter
- Vivarium (7) Apply Vivarium filter
Publication Date
- 2025 (85) Apply 2025 filter
- 2024 (221) Apply 2024 filter
- 2023 (160) Apply 2023 filter
- 2022 (167) Apply 2022 filter
- 2021 (175) Apply 2021 filter
- 2020 (177) Apply 2020 filter
- 2019 (177) Apply 2019 filter
- 2018 (206) Apply 2018 filter
- 2017 (186) Apply 2017 filter
- 2016 (191) Apply 2016 filter
- 2015 (195) Apply 2015 filter
- 2014 (190) Apply 2014 filter
- 2013 (136) Apply 2013 filter
- 2012 (112) Apply 2012 filter
- 2011 (98) Apply 2011 filter
- 2010 (61) Apply 2010 filter
- 2009 (56) Apply 2009 filter
- 2008 (40) Apply 2008 filter
- 2007 (21) Apply 2007 filter
- 2006 (3) Apply 2006 filter
2657 Janelia Publications
Showing 1831-1840 of 2657 resultsThe hippocampus exhibits a variety of distinct states of activity under different conditions. For instance the rhythmic patterns of activity orchestrated by the theta oscillation during running and REM sleep are markedly different from the large irregular activity (LIA) observed during awake resting and slow wave sleep. We found that under different levels of isoflurane anesthesia activity in the hippocampus of rats displays two distinct states which have several qualities that mirror the theta and LIA states. These data provide further evidence that the two states are intrinsic modes of the hippocampus; while also characterizing a preparation that could be useful for studying the natural activity states in hippocampus. This article is protected by copyright. All rights reserved.
A striking aspect of cortical neural networks is the divergence of a relatively small number of input channels from the peripheral sensory apparatus into a large number of cortical neurons, an over-complete representation strategy. Cortical neurons are then connected by a sparse network of lateral synapses. Here we propose that such architecture may increase the persistence of the representation of an incoming stimulus, or a percept. We demonstrate that for a family of networks in which the receptive field of each neuron is re-expressed by its outgoing connections, a represented percept can remain constant despite changing activity. We term this choice of connectivity REceptive FIeld REcombination (REFIRE) networks. The sparse REFIRE network may serve as a high-dimensional integrator and a biologically plausible model of the local cortical circuit.
Electron cryomicroscopy, or cryoEM, is an emerging technique for studying the three-dimensional structures of proteins and large macromolecular machines. Electron crystallography is a branch of cryoEM in which structures of proteins can be studied at resolutions that rival those achieved by X-ray crystallography. Electron crystallography employs two-dimensional crystals of a membrane protein embedded within a lipid bilayer. The key to a successful electron crystallographic experiment is the crystallization, or reconstitution, of the protein of interest. This unit describes ways in which protein can be expressed, purified, and reconstituted into well-ordered two-dimensional crystals. A protocol is also provided for negative stain electron microscopy as a tool for screening crystallization trials. When large and well-ordered crystals are obtained, the structures of both protein and its surrounding membrane can be determined to atomic resolution.
How brains are hardwired to produce aggressive behavior, and how aggression circuits are related to those that mediate courtship, is not well understood. A large-scale screen for aggression-promoting neurons in Drosophila identified several independent hits that enhanced both inter-male aggression and courtship. Genetic intersections revealed that 8-10 P1 interneurons, previously thought to exclusively control male courtship, were sufficient to promote fighting. Optogenetic experiments indicated that P1 activation could promote aggression at a threshold below that required for wing extension. P1 activation in the absence of wing extension triggered persistent aggression via an internal state that could endure for minutes. High-frequency P1 activation promoted wing extension and suppressed aggression during photostimulation, whereas aggression resumed and wing extension was inhibited following photostimulation offset. Thus, P1 neuron activation promotes a latent, internal state that facilitates aggression and courtship, and controls the overt expression of these social behaviors in a threshold-dependent, inverse manner.
Live-cell fluorescence light microscopy has emerged as an important tool in the study of cellular biology. The development of fluorescent markers in parallel with super-resolution imaging systems has pushed light microscopy into the realm of molecular visualization at the nanometer scale. Resolutions previously only attained with electron microscopes are now within the grasp of light microscopes. However, until recently, live-cell imaging approaches have eluded super-resolution microscopy, hampering it from reaching its full potential for revealing the dynamic interactions in biology occurring at the single molecule level. Here we examine recent advances in the super-resolution imaging of living cells by reviewing recent breakthroughs in single molecule localization microscopy methods such as PALM and STORM to achieve this important goal.
Ultrasound pulse guided digital phase conjugation has emerged to realize fluorescence imaging inside random scattering media. Its major limitation is the slow imaging speed, as a new wavefront needs to be measured for each voxel. Therefore 3D or even 2D imaging can be time consuming. For practical applications on biological systems, we need to accelerate the imaging process by orders of magnitude. Here we propose and experimentally demonstrate a parallel wavefront measurement scheme towards such a goal. Multiple focused ultrasound pulses of different carrier frequencies can be simultaneously launched inside a scattering medium. Heterodyne interferometry is used to measure all of the wavefronts originating from every sound focus in parallel. We use these wavefronts in sequence to rapidly excite fluorescence at all the voxels defined by the focused ultrasound pulses. In this report, we employed a commercially available sound transducer to generate two sound foci in parallel, doubled the wavefront measurement speed, and reduced the mechanical scanning steps of the sound transducer to half.
A parallel wavefront optimization method is demonstrated experimentally to focus light through random scattering media. The simultaneous modulation of multiple phase elements, each at a unique frequency, enables a parallel determination of the optimal wavefront. Compared to a pixel-by-pixel measurement, the reported parallel method uses the target signal in a highly efficient way. With 441 phase elements, a high-quality focus was formed through a glass diffuser with a peak-to-background ratio of \~{}270. The accuracy and repeatability of the system were tested through experiments.
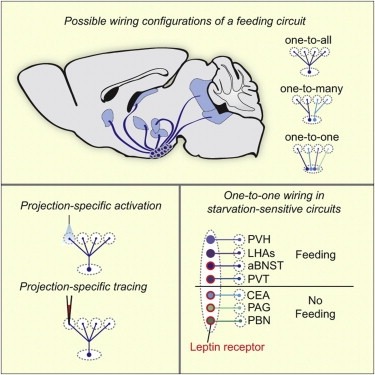
Neural circuits for essential natural behaviors are shaped by selective pressure to coordinate reliable execution of flexible goal-directed actions. However, the structural and functional organization of survival-oriented circuits is poorly understood due to exceptionally complex neuroanatomy. This is exemplified by AGRP neurons, which are a molecularly defined population that is sufficient to rapidly coordinate voracious food seeking and consumption behaviors. Here, we use cell-type-specific techniques for neural circuit manipulation and projection-specific anatomical analysis to examine the organization of this critical homeostatic circuit that regulates feeding. We show that AGRP neuronal circuits use a segregated, parallel, and redundant output configuration. AGRP neuron axon projections that target different brain regions originate from distinct subpopulations, several of which are sufficient to independently evoke feeding. The concerted anatomical and functional analysis of AGRP neuron projection populations reveals a constellation of core forebrain nodes, which are part of an extended circuit that mediates feeding behavior.