Filter
Associated Lab
- Aguilera Castrejon Lab (1) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (53) Apply Ahrens Lab filter
- Aso Lab (40) Apply Aso Lab filter
- Baker Lab (19) Apply Baker Lab filter
- Betzig Lab (101) Apply Betzig Lab filter
- Beyene Lab (8) Apply Beyene Lab filter
- Bock Lab (14) Apply Bock Lab filter
- Branson Lab (50) Apply Branson Lab filter
- Card Lab (36) Apply Card Lab filter
- Cardona Lab (45) Apply Cardona Lab filter
- Chklovskii Lab (10) Apply Chklovskii Lab filter
- Clapham Lab (14) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (8) Apply Darshan Lab filter
- Dickson Lab (32) Apply Dickson Lab filter
- Druckmann Lab (21) Apply Druckmann Lab filter
- Dudman Lab (38) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (4) Apply Egnor Lab filter
- Espinosa Medina Lab (15) Apply Espinosa Medina Lab filter
- Feliciano Lab (7) Apply Feliciano Lab filter
- Fetter Lab (31) Apply Fetter Lab filter
- Fitzgerald Lab (16) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (38) Apply Funke Lab filter
- Gonen Lab (59) Apply Gonen Lab filter
- Grigorieff Lab (34) Apply Grigorieff Lab filter
- Harris Lab (53) Apply Harris Lab filter
- Heberlein Lab (13) Apply Heberlein Lab filter
- Hermundstad Lab (22) Apply Hermundstad Lab filter
- Hess Lab (74) Apply Hess Lab filter
- Ilanges Lab (2) Apply Ilanges Lab filter
- Jayaraman Lab (42) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (1) Apply Johnson Lab filter
- Karpova Lab (13) Apply Karpova Lab filter
- Keleman Lab (8) Apply Keleman Lab filter
- Keller Lab (61) Apply Keller Lab filter
- Koay Lab (2) Apply Koay Lab filter
- Lavis Lab (137) Apply Lavis Lab filter
- Lee (Albert) Lab (29) Apply Lee (Albert) Lab filter
- Leonardo Lab (19) Apply Leonardo Lab filter
- Li Lab (4) Apply Li Lab filter
- Lippincott-Schwartz Lab (97) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (1) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (58) Apply Liu (Zhe) Lab filter
- Looger Lab (137) Apply Looger Lab filter
- Magee Lab (31) Apply Magee Lab filter
- Menon Lab (12) Apply Menon Lab filter
- Murphy Lab (6) Apply Murphy Lab filter
- O'Shea Lab (6) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Pachitariu Lab (36) Apply Pachitariu Lab filter
- Pastalkova Lab (5) Apply Pastalkova Lab filter
- Pavlopoulos Lab (7) Apply Pavlopoulos Lab filter
- Pedram Lab (4) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (45) Apply Reiser Lab filter
- Riddiford Lab (20) Apply Riddiford Lab filter
- Romani Lab (31) Apply Romani Lab filter
- Rubin Lab (105) Apply Rubin Lab filter
- Saalfeld Lab (46) Apply Saalfeld Lab filter
- Satou Lab (1) Apply Satou Lab filter
- Scheffer Lab (36) Apply Scheffer Lab filter
- Schreiter Lab (50) Apply Schreiter Lab filter
- Sgro Lab (1) Apply Sgro Lab filter
- Shroff Lab (31) Apply Shroff Lab filter
- Simpson Lab (18) Apply Simpson Lab filter
- Singer Lab (37) Apply Singer Lab filter
- Spruston Lab (57) Apply Spruston Lab filter
- Stern Lab (73) Apply Stern Lab filter
- Sternson Lab (47) Apply Sternson Lab filter
- Stringer Lab (32) Apply Stringer Lab filter
- Svoboda Lab (131) Apply Svoboda Lab filter
- Tebo Lab (9) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (18) Apply Tillberg Lab filter
- Tjian Lab (17) Apply Tjian Lab filter
- Truman Lab (58) Apply Truman Lab filter
- Turaga Lab (39) Apply Turaga Lab filter
- Turner Lab (27) Apply Turner Lab filter
- Vale Lab (7) Apply Vale Lab filter
- Voigts Lab (3) Apply Voigts Lab filter
- Wang (Meng) Lab (21) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (6) Apply Wang (Shaohe) Lab filter
- Wu Lab (8) Apply Wu Lab filter
- Zlatic Lab (26) Apply Zlatic Lab filter
- Zuker Lab (5) Apply Zuker Lab filter
Associated Project Team
- CellMap (12) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- FIB-SEM Technology (3) Apply FIB-SEM Technology filter
- Fly Descending Interneuron (11) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (53) Apply FlyEM filter
- FlyLight (49) Apply FlyLight filter
- GENIE (46) Apply GENIE filter
- Integrative Imaging (4) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (18) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (26) Apply Tool Translation Team (T3) filter
- Transcription Imaging (45) Apply Transcription Imaging filter
Associated Support Team
- Project Pipeline Support (5) Apply Project Pipeline Support filter
- Anatomy and Histology (18) Apply Anatomy and Histology filter
- Cryo-Electron Microscopy (35) Apply Cryo-Electron Microscopy filter
- Electron Microscopy (16) Apply Electron Microscopy filter
- Gene Targeting and Transgenics (11) Apply Gene Targeting and Transgenics filter
- Integrative Imaging (17) Apply Integrative Imaging filter
- Invertebrate Shared Resource (40) Apply Invertebrate Shared Resource filter
- Janelia Experimental Technology (37) Apply Janelia Experimental Technology filter
- Management Team (1) Apply Management Team filter
- Molecular Genomics (15) Apply Molecular Genomics filter
- Primary & iPS Cell Culture (14) Apply Primary & iPS Cell Culture filter
- Project Technical Resources (50) Apply Project Technical Resources filter
- Quantitative Genomics (19) Apply Quantitative Genomics filter
- Scientific Computing Software (92) Apply Scientific Computing Software filter
- Scientific Computing Systems (7) Apply Scientific Computing Systems filter
- Viral Tools (14) Apply Viral Tools filter
- Vivarium (7) Apply Vivarium filter
Publication Date
- 2025 (124) Apply 2025 filter
- 2024 (215) Apply 2024 filter
- 2023 (159) Apply 2023 filter
- 2022 (167) Apply 2022 filter
- 2021 (175) Apply 2021 filter
- 2020 (177) Apply 2020 filter
- 2019 (177) Apply 2019 filter
- 2018 (206) Apply 2018 filter
- 2017 (186) Apply 2017 filter
- 2016 (191) Apply 2016 filter
- 2015 (195) Apply 2015 filter
- 2014 (190) Apply 2014 filter
- 2013 (136) Apply 2013 filter
- 2012 (112) Apply 2012 filter
- 2011 (98) Apply 2011 filter
- 2010 (61) Apply 2010 filter
- 2009 (56) Apply 2009 filter
- 2008 (40) Apply 2008 filter
- 2007 (21) Apply 2007 filter
- 2006 (3) Apply 2006 filter
2689 Janelia Publications
Showing 2511-2520 of 2689 resultsDrosophila show innate olfactory-driven behaviours that are observed in naive animals without previous learning or experience, suggesting that the neural circuits that mediate these behaviours are genetically programmed. Despite the numerical simplicity of the fly nervous system, features of the anatomical organization of the fly brain often confound the delineation of these circuits. Here we identify a neural circuit responsive to cVA, a pheromone that elicits sexually dimorphic behaviours. We have combined neural tracing using an improved photoactivatable green fluorescent protein (PA-GFP) with electrophysiology, optical imaging and laser-mediated microlesioning to map this circuit from the activation of sensory neurons in the antennae to the excitation of descending neurons in the ventral nerve cord. This circuit is concise and minimally comprises four neurons, connected by three synapses. Three of these neurons are overtly dimorphic and identify a male-specific neuropil that integrates inputs from multiple sensory systems and sends outputs to the ventral nerve cord. This neural pathway suggests a means by which a single pheromone can elicit different behaviours in the two sexes.
Cell adhesions to the extracellular matrix (ECM) are necessary for morphogenesis, immunity, and wound healing. Focal adhesions are multifunctional organelles that mediate cell-ECM adhesion, force transmission, cytoskeletal regulation and signaling. Focal adhesions consist of a complex network of trans-plasma-membrane integrins and cytoplasmic proteins that form a <200-nm plaque linking the ECM to the actin cytoskeleton. The complexity of focal adhesion composition and dynamics implicate an intricate molecular machine. However, focal adhesion molecular architecture remains unknown. Here we used three-dimensional super-resolution fluorescence microscopy (interferometric photoactivated localization microscopy) to map nanoscale protein organization in focal adhesions. Our results reveal that integrins and actin are vertically separated by a \~{}40-nm focal adhesion core region consisting of multiple protein-specific strata: a membrane-apposed integrin signaling layer containing integrin cytoplasmic tails, focal adhesion kinase, and paxillin; an intermediate force-transduction layer containing talin and vinculin; and an uppermost actin-regulatory layer containing zyxin, vasodilator-stimulated phosphoprotein and α-actinin. By localizing amino- and carboxy-terminally tagged talins, we reveal talin’s polarized orientation, indicative of a role in organizing the focal adhesion strata. The composite multilaminar protein architecture provides a molecular blueprint for understanding focal adhesion functions.
This mini-symposium aims to provide an integrated perspective on recent developments in optogenetics. Research in this emerging field combines optical methods with targeted expression of genetically encoded, protein-based probes to achieve experimental manipulation and measurement of neural systems with superior temporal and spatial resolution. The essential components of the optogenetic toolbox consist of two kinds of molecular devices: actuators and reporters, which respectively enable light-mediated control or monitoring of molecular processes. The first generation of genetically encoded calcium reporters, fluorescent proteins, and neural activators has already had a great impact on neuroscience. Now, a second generation of voltage reporters, neural silencers, and functionally extended fluorescent proteins hold great promise for continuing this revolution. In this review, we will evaluate and highlight the limitations of presently available optogenic tools and discuss where these technologies and their applications are headed in the future.
Spatial navigation is often used as a behavioral task in studies of the neuronal circuits that underlie cognition, learning and memory in rodents. The combination of in vivo microscopy with genetically encoded indicators has provided an important new tool for studying neuronal circuits, but has been technically difficult to apply during navigation. Here we describe methods for imaging the activity of neurons in the CA1 region of the hippocampus with subcellular resolution in behaving mice. Neurons that expressed the genetically encoded calcium indicator GCaMP3 were imaged through a chronic hippocampal window. Head-restrained mice performed spatial behaviors in a setup combining a virtual reality system and a custom-built two-photon microscope. We optically identified populations of place cells and determined the correlation between the location of their place fields in the virtual environment and their anatomical location in the local circuit. The combination of virtual reality and high-resolution functional imaging should allow a new generation of studies to investigate neuronal circuit dynamics during behavior.
The primary auditory cortex (A1) is organized tonotopically, with neurons sensitive to high and low frequencies arranged in a rostro-caudal gradient. We used laser scanning photostimulation in acute slices to study the organization of local excitatory connections onto layers 2 and 3 (L2/3) of the mouse A1. Consistent with the organization of other cortical regions, synaptic inputs along the isofrequency axis (orthogonal to the tonotopic axis) arose predominantly within a column. By contrast, we found that local connections along the tonotopic axis differed from those along the isofrequency axis: some input pathways to L3 (but not L2) arose predominantly out-of-column. In vivo cell-attached recordings revealed differences between the sound-responsiveness of neurons in L2 and L3. Our results are consistent with the hypothesis that auditory cortical microcircuitry is specialized to the one-dimensional representation of frequency in the auditory cortex.
The optic tectum of zebrafish is involved in behavioral responses that require the detection of small objects. The superficial layers of the tectal neuropil receive input from retinal axons, while its deeper layers convey the processed information to premotor areas. Imaging with a genetically encoded calcium indicator revealed that the deep layers, as well as the dendrites of single tectal neurons, are preferentially activated by small visual stimuli. This spatial filtering relies on GABAergic interneurons (using the neurotransmitter γ-aminobutyric acid) that are located in the superficial input layer and respond only to large visual stimuli. Photo-ablation of these cells with KillerRed, or silencing of their synaptic transmission, eliminates the size tuning of deeper layers and impairs the capture of prey.
This paper provides a compilation of diagrammatic representations of the expression profiles of epidermal and fat body mRNAs during the last two larval instars and metamorphosis of the tobacco hornworm, Manduca sexta. Included are those encoding insecticyanin, three larval cuticular proteins, dopa decarboxylase, moling, and the juvenile hormone-binding protein JP29 produced by the dorsal abdominal epidermis, and arylphorin and the methionine-rich storage proteins made by the fat body. The mRNA profiles of the ecdysteroid-regulated cascade of transcription factors in the epidermis during the larval molt and the onset of metamorphosis and in the pupal wing during the onset of adult development are also shown. These profiles are accompanied by a brief summary of the current knowledge about the regulation of these mRNAs by ecdysteroids and juvenile hormone based on experimental manipulations, both in vivo and in vitro.
The rapid adoption of high-throughput next generation sequence data in biological research is presenting a major challenge for sequence alignment tools—specifically, the efficient alignment of vast amounts of short reads to large references in the presence of differences arising from sequencing errors and biological sequence variations. To address this challenge, we developed a short read aligner for high-throughput sequencer data that is tolerant of errors or mutations of all types—namely, substitutions, deletions, and insertions. The aligner utilizes a multi-stage approach in which template-based indexing is used to identify candidate regions for alignment with dynamic programming. A template is a pair of gapped seeds, with one used with the read and one used with the reference. In this article, we focus on the development of template families that yield error-tolerant indexing up to a given error-budget. A general algorithm for finding those families is presented, and a recursive construction that creates families with higher error tolerance from ones with a lower error tolerance is developed.
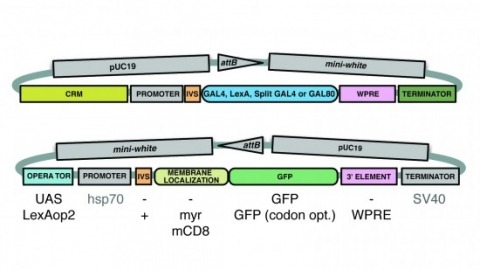
A wide variety of biological experiments rely on the ability to express an exogenous gene in a transgenic animal at a defined level and in a spatially and temporally controlled pattern. We describe major improvements of the methods available for achieving this objective in Drosophila melanogaster. We have systematically varied core promoters, UTRs, operator sequences, and transcriptional activating domains used to direct gene expression with the GAL4, LexA, and Split GAL4 transcription factors and the GAL80 transcriptional repressor. The use of site-specific integration allowed us to make quantitative comparisons between different constructs inserted at the same genomic location. We also characterized a set of PhiC31 integration sites for their ability to support transgene expression of both drivers and responders in the nervous system. The increased strength and reliability of these optimized reagents overcome many of the previous limitations of these methods and will facilitate genetic manipulations of greater complexity and sophistication.
Reconstructing neuronal circuits at the level of synapses is a central problem in neuroscience, and the focus of the nascent field of connectomics. Previously used to reconstruct the C. elegans wiring diagram, serial-section transmission electron microscopy (ssTEM) is a proven technique for the task. However, to reconstruct more complex circuits, ssTEM will require the automation of image processing. We review progress in the processing of electron microscopy images and, in particular, a semi-automated reconstruction pipeline deployed at Janelia. Drosophila circuits underlying identified behaviors are being reconstructed in the pipeline with the goal of generating a complete Drosophila connectome.