Select Publications
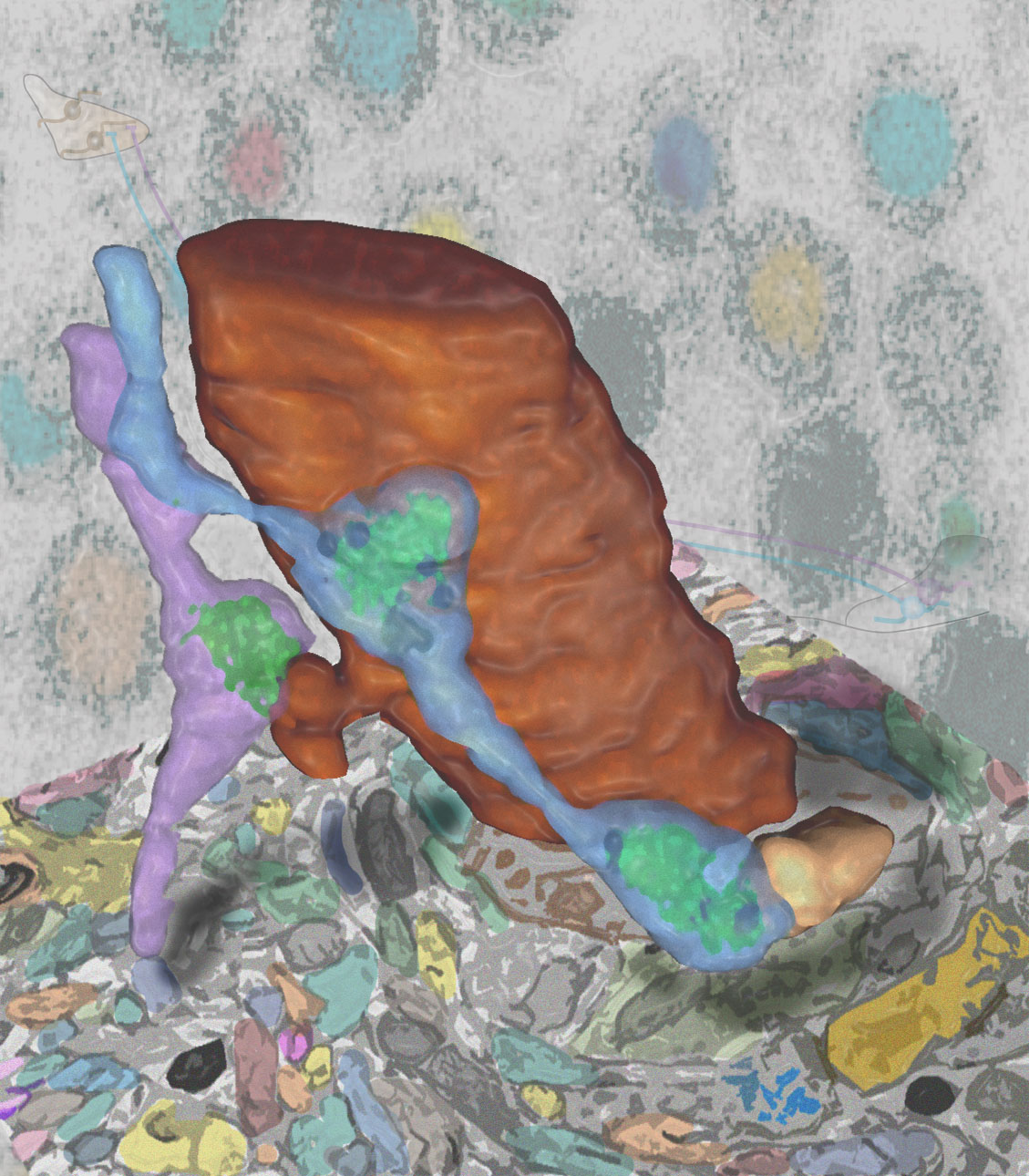
Natural events present multiple types of sensory cues, each detected by a specialized sensory modality. Combining information from several modalities is essential for the selection of appropriate actions. Key to understanding multimodal computations is determining the structural patterns of multimodal convergence and how these patterns contribute to behaviour. Modalities could converge early, late or at multiple levels in the sensory processing hierarchy. Here we show that combining mechanosensory and nociceptive cues synergistically enhances the selection of the fastest mode of escape locomotion in Drosophila larvae. In an electron microscopy volume that spans the entire insect nervous system, we reconstructed the multisensory circuit supporting the synergy, spanning multiple levels of the sensory processing hierarchy. The wiring diagram revealed a complex multilevel multimodal convergence architecture. Using behavioural and physiological studies, we identified functionally connected circuit nodes that trigger the fastest locomotor mode, and others that facilitate it, and we provide evidence that multiple levels of multimodal integration contribute to escape mode selection. We propose that the multilevel multimodal convergence architecture may be a general feature of multisensory circuits enabling complex input–output functions and selective tuning to ecologically relevant combinations of cues.
View Publication PageTransgenesis in numerous eukaryotes has been facilitated by the use of site-specific integrases to stably insert transgenes at predefined genomic positions (landing sites). However, the utility of integrase-mediated transgenesis in any system is constrained by the limited number and variable expression properties of available landing sites. By exploiting the nonstandard recombination activity exhibited by a phiC31 integrase mutant, we developed a rapid and inexpensive method for isolating landing sites that exhibit desired expression properties. Additionally, we devised a simple technique for constructing arrays of transgenes at a single landing site, thereby extending the utility of previously characterized landing sites. Using the fruit fly Drosophila melanogaster, we demonstrate the feasibility of these approaches by isolating new landing sites optimized to express transgenes in the nervous system and by building fluorescent reporter arrays at several landing sites. Because these strategies require the activity of only a single exogenous protein, we anticipate that they will be portable to species such as nonmodel organisms, in which genetic manipulation is more challenging, expediting the development of genetic resources in these systems.
View Publication PageSynaptic connectivity and molecular composition provide a blueprint for information processing in neural circuits. Detailed structural analysis of neural circuits requires nanometer resolution, which can be obtained with serial-section electron microscopy. However, this technique remains challenging for reconstructing molecularly defined synapses. We used a genetically encoded synaptic marker for electron microscopy (GESEM) based on intra-vesicular generation of electron-dense labeling in axonal boutons. This approach allowed the identification of synapses from Cre recombinase-expressing or GAL4-expressing neurons in the mouse and fly with excellent preservation of ultrastructure. We applied this tool to visualize long-range connectivity of AGRP and POMC neurons in the mouse, two molecularly defined hypothalamic populations that are important for feeding behavior. Combining selective ultrastructural reconstruction of neuropil with functional and viral circuit mapping, we characterized some basic features of circuit organization for axon projections of these cell types. Our findings demonstrate that GESEM labeling enables long-range connectomics with molecularly defined cell types.
View Publication PageMotor sequences are formed through the serial execution of different movements, but how nervous systems implement this process remains largely unknown. We determined the organizational principles governing how dirty fruit flies groom their bodies with sequential movements. Using genetically targeted activation of neural subsets, we drove distinct motor programs that clean individual body parts. This enabled competition experiments revealing that the motor programs are organized into a suppression hierarchy; motor programs that occur first suppress those that occur later. Cleaning one body part reduces the sensory drive to its motor program, which relieves suppression of the next movement, allowing the grooming sequence to progress down the hierarchy. A model featuring independently evoked cleaning movements activated in parallel, but selected serially through hierarchical suppression, was successful in reproducing the grooming sequence. This provides the first example of an innate motor sequence implemented by the prevailing model for generating human action sequences.
View Publication PageDespite the importance of the insect nervous system for functional and developmental neuroscience, descriptions of insect brains have suffered from a lack of uniform nomenclature. Ambiguous definitions of brain regions and fiber bundles have contributed to the variation of names used to describe the same structure. The lack of clearly determined neuropil boundaries has made it difficult to document precise locations of neuronal projections for connectomics study. To address such issues, a consortium of neurobiologists studying arthropod brains, the Insect Brain Name Working Group, has established the present hierarchical nomenclature system, using the brain of Drosophila melanogaster as the reference framework, while taking the brains of other taxa into careful consideration for maximum consistency and expandability. The following summarizes the consortium’s nomenclature system and highlights examples of existing ambiguities and remedies for them. This nomenclature is intended to serve as a standard of reference for the study of the brain of Drosophila and other insects.
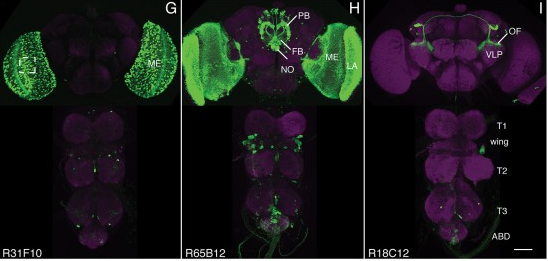
View Publication PageWe established a collection of 7,000 transgenic lines of Drosophila melanogaster. Expression of GAL4 in each line is controlled by a different, defined fragment of genomic DNA that serves as a transcriptional enhancer. We used confocal microscopy of dissected nervous systems to determine the expression patterns driven by each fragment in the adult brain and ventral nerve cord. We present image data on 6,650 lines. Using both manual and machine-assisted annotation, we describe the expression patterns in the most useful lines. We illustrate the utility of these data for identifying novel neuronal cell types, revealing brain asymmetry, and describing the nature and extent of neuronal shape stereotypy. The GAL4 lines allow expression of exogenous genes in distinct, small subsets of the adult nervous system. The set of DNA fragments, each driving a documented expression pattern, will facilitate the generation of additional constructs for manipulating neuronal function. synapse was substantially elevated, at the endocytic zone there was no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
For the overall strategy and methods used to produce the GAL4 lines:
Pfeiffer, B.D., Jenett, A., Hammonds, A.S., Ngo, T.T., Misra, S., Murphy, C., Scully, A., Carlson, J.W., Wan, K.H., Laverty, T.R., Mungall, C., Svirskas, R., Kadonaga, J.T., Doe, C.Q., Eisen, M.B., Celniker, S.E., Rubin, G.M. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715-9720. http://www.pnas.org/content/105/28/9715.full.pdf+html synapse was substantially elevated, at the endocytic zone there was no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
For data on expression in the embryo:
Manning, L., Purice, M.D., Roberts, J., Pollard, J.L., Bennett, A.L., Kroll, J.R., Dyukareva, A.V., Doan, P.N., Lupton, J.R., Strader, M.E., Tanner, S., Bauer, D., Wilbur, A., Tran, K.D., Laverty, T.R., Pearson, J.C., Crews, S.T., Rubin, G.M., and Doe, C.Q. (2012) Annotated embryonic CNS expression patterns of 5000 GMR GAL4 lines: a resource for manipulating gene expression and analyzing cis-regulatory motifs. Cell Reports (2012) Doi: 10.1016/j.celrep.2012.09.009 http://www.cell.com/cell-reports/fulltext/S2211-1247(12)00290-2 synapse was substantially elevated, at the endocytic zone there was no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
For data on expression in imaginal discs:
Jory, A., Estella, C., Giorgianni, M.W., Slattery, M., Laverty, T.R., Rubin, G.M., and Mann, R.S. (2012) A survey of 6300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophila melanogaster. Cell Reports (2012) Doi: 10.1016/j.celrep.2012.09.010 http://www.cell.com/cell-reports/fulltext/S2211-1247(12)00291-4 synapse was substantially elevated, at the endocytic zone there was no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
For data on expression in the larval nervous system:
Li, H.-H., Kroll, J.R., Lennox, S., Ogundeyi, O., Jeter, J., Depasquale, G., and Truman, J.W. (2013) A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Reports (submitted).
Analyzing Drosophila melanogaster neural expression patterns in thousands of three-dimensional image stacks of individual brains requires registering them into a canonical framework based on a fiducial reference of neuropil morphology. Given a target brain labeled with predefined landmarks, the BrainAligner program automatically finds the corresponding landmarks in a subject brain and maps it to the coordinate system of the target brain via a deformable warp. Using a neuropil marker (the antibody nc82) as a reference of the brain morphology and a target brain that is itself a statistical average of data for 295 brains, we achieved a registration accuracy of 2 μm on average, permitting assessment of stereotypy, potential connectivity and functional mapping of the adult fruit fly brain. We used BrainAligner to generate an image pattern atlas of 2954 registered brains containing 470 different expression patterns that cover all the major compartments of the fly brain.
View Publication PageWe developed a multicolor neuron labeling technique in Drosophila melanogaster that combines the power to specifically target different neural populations with the label diversity provided by stochastic color choice. This adaptation of vertebrate Brainbow uses recombination to select one of three epitope-tagged proteins detectable by immunofluorescence. Two copies of this construct yield six bright, separable colors. We used Drosophila Brainbow to study the innervation patterns of multiple antennal lobe projection neuron lineages in the same preparation and to observe the relative trajectories of individual aminergic neurons. Nerve bundles, and even individual neurites hundreds of micrometers long, can be followed with definitive color labeling. We traced motor neurons in the subesophageal ganglion and correlated them to neuromuscular junctions to identify their specific proboscis muscle targets. The ability to independently visualize multiple lineage or neuron projections in the same preparation greatly advances the goal of mapping how neurons connect into circuits.
View Publication PageThe role of gamma amino butyric acid (GABA) release and inhibitory neurotransmission in regulating most behaviors remains unclear. The vesicular GABA transporter (VGAT) is required for the storage of GABA in synaptic vesicles and provides a potentially useful probe for inhibitory circuits. However, specific pharmacologic agents for VGAT are not available, and VGAT knockout mice are embryonically lethal, thus precluding behavioral studies. We have identified the Drosophila ortholog of the vesicular GABA transporter gene (which we refer to as dVGAT), immunocytologically mapped dVGAT protein expression in the larva and adult and characterized a dVGAT(minos) mutant allele. dVGAT is embryonically lethal and we do not detect residual dVGAT expression, suggesting that it is either a strong hypomorph or a null. To investigate the function of VGAT and GABA signaling in adult visual flight behavior, we have selectively rescued the dVGAT mutant during development. We show that reduced GABA release does not compromise the active optomotor control of wide-field pattern motion. Conversely, reduced dVGAT expression disrupts normal object tracking and figure-ground discrimination. These results demonstrate that visual behaviors are segregated by the level of GABA signaling in flies, and more generally establish dVGAT as a model to study the contribution of GABA release to other complex behaviors.
View Publication PageThe V3D system provides three-dimensional (3D) visualization of gigabyte-sized microscopy image stacks in real time on current laptops and desktops. V3D streamlines the online analysis, measurement and proofreading of complicated image patterns by combining ergonomic functions for selecting a location in an image directly in 3D space and for displaying biological measurements, such as from fluorescent probes, using the overlaid surface objects. V3D runs on all major computer platforms and can be enhanced by software plug-ins to address specific biological problems. To demonstrate this extensibility, we built a V3D-based application, V3D-Neuron, to reconstruct complex 3D neuronal structures from high-resolution brain images. V3D-Neuron can precisely digitize the morphology of a single neuron in a fruitfly brain in minutes, with about a 17-fold improvement in reliability and tenfold savings in time compared with other neuron reconstruction tools. Using V3D-Neuron, we demonstrate the feasibility of building a 3D digital atlas of neurite tracts in the fruitfly brain.
View Publication PageAutomatic alignment (registration) of 3D images of adult fruit fly brains is often influenced by the significant displacement of the relative locations of the two optic lobes (OLs) and the center brain (CB). In one of our ongoing efforts to produce a better image alignment pipeline of adult fruit fly brains, we consider separating CB and OLs and align them independently. This paper reports our automatic method to segregate CB and OLs, in particular under conditions where the signal to noise ratio (SNR) is low, the variation of the image intensity is big, and the relative displacement of OLs and CB is substantial. We design an algorithm to find a minimum-cost 3D surface in a 3D image stack to best separate an OL (of one side, either left or right) from CB. This surface is defined as an aggregation of the respective minimum-cost curves detected in each individual 2D image slice. Each curve is defined by a list of control points that best segregate OL and CB. To obtain the locations of these control points, we derive an energy function that includes an image energy term defined by local pixel intensities and two internal energy terms that constrain the curve’s smoothness and length. Gradient descent method is used to optimize this energy function. To improve both the speed and robustness of the method, for each stack, the locations of optimized control points in a slice are taken as the initialization prior for the next slice. We have tested this approach on simulated and real 3D fly brain image stacks and demonstrated that this method can reasonably segregate OLs from CBs despite the aforementioned difficulties.
View Publication PageDrosophila is a marvelous system to study the underlying principles that govern how neural circuits govern behaviors. The scale of the fly brain (approximately 100,000 neurons) and the complexity of the behaviors the fly can perform make it a tractable experimental model organism. In addition, 100 years and hundreds of labs have contributed to an extensive array of tools and techniques that can be used to dissect the function and organization of the fly nervous system. This review discusses both the conceptual challenges and the specific tools for a neurogenetic approach to circuit mapping in Drosophila.
View Publication PageNF-κB signaling has been implicated in neurodegenerative disease, epilepsy, and neuronal plasticity. However, the cellular and molecular activity of NF-κB signaling within the nervous system remains to be clearly defined. Here, we show that the NF-κB and IκB homologs Dorsal and Cactus surround postsynaptic glutamate receptor (GluR) clusters at the Drosophila NMJ. We then show that mutations in dorsal, cactus, and IRAK/pelle kinase specifically impair GluR levels, assayed immunohistochemically and electrophysiologically, without affecting NMJ growth, the size of the postsynaptic density, or homeostatic plasticity. Additional genetic experiments support the conclusion that cactus functions in concert with, rather than in opposition to, dorsal and pelle in this process. Finally, we provide evidence that Dorsal and Cactus act posttranscriptionally, outside the nucleus, to control GluR density. Based upon our data we speculate that Dorsal, Cactus, and Pelle could function together, locally at the postsynaptic density, to specify GluR levels.
View Publication PageThe Roundabout (Robo) receptors have been intensively studied for their role in regulating axon guidance in the embryonic nervous system, whereas a role in dendritic guidance has not been explored. In the adult giant fiber system of Drosophila, we have revealed that ectopic Robo expression can regulate the growth and guidance of specific motor neuron dendrites, whereas Robo2 and Robo3 have no effect. We also show that the effect of Robo on dendritic guidance can be suppressed by Commissureless coexpression. Although we confirmed a role for all three Robo receptors in giant fiber axon guidance, the strong axon guidance alterations caused by overexpression of Robo2 or Robo3 have no effect on synaptic connectivity. In contrast, Robo overexpression in the giant fiber seems to directly interfere with synaptic function. We conclude that axon guidance, dendritic guidance, and synaptogenesis are separable processes and that the different Robo family members affect them distinctly.
View Publication PageSlit is secreted by cells at the midline of the central nervous system, where it binds to Roundabout (Robo) receptors and functions as a potent repellent. We found that migrating mesodermal cells in vivo respond to Slit as both an attractant and a repellent and that Robo receptors are required for both functions. Mesoderm cells expressing Robo receptors initially migrate away from Slit at the midline. A few hours after migration, these same cells change their behavior and require Robo to extend toward Slit-expressing muscle attachment sites. Thus, Slit functions as a chemoattractant to provide specificity for muscle patterning.
View Publication PageSlit is secreted by midline glia in Drosophila and functions as a short-range repellent to control midline crossing. Although most Slit stays near the midline, some diffuses laterally, functioning as a long-range chemorepellent. Here we show that a combinatorial code of Robo receptors controls lateral position in the CNS by responding to this presumptive Slit gradient. Medial axons express only Robo, intermediate axons express Robo3 and Robo, while lateral axons express Robo2, Robo3, and Robo. Removal of robo2 or robo3 causes lateral axons to extend medially; ectopic expression of Robo2 or Robo3 on medial axons drives them laterally. Precise topography of longitudinal pathways appears to be controlled by a combination of long-range guidance (the Robo code determining region) and short-range guidance (discrete local cues determining specific location within a region).
View Publication PagePrevious studies showed that Roundabout (Robo) in Drosophila is a repulsive axon guidance receptor that binds to Slit, a repellent secreted by midline glia. In robo mutants, growth cones cross and recross the midline, while, in slit mutants, growth cones enter the midline but fail to leave it. This difference suggests that Slit must have more than one receptor controlling midline guidance. In the absence of Robo, some other Slit receptor ensures that growth cones do not stay at the midline, even though they cross and recross it. Here we show that the Drosophila genome encodes three Robo receptors and that Robo and Robo2 have distinct functions, which together control repulsive axon guidance at the midline. The robo,robo2 double mutant is largely identical to slit.
View Publication Page