 The "Stitcher" is a suite of programs for registering terabytes of transmission electron microscopy (ssTEM) and optical microscopy data.
The "Stitcher" is a suite of programs for registering terabytes of transmission electron microscopy (ssTEM) and optical microscopy data.
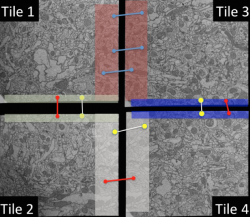
ssTEM imaging is used in elucidating the anatomical connectivity among neurons at the scale of axons, dendrites and synapses in the context of the whole brain. The technique generates images (image tiles) that must be assembled in a post-acquisition step to reconstruct the full image volume.
The physical coordinates provided by the microscope are not precise enough to directly allow such assembly from individual image tiles. Staining artifacts, mechanical deformation, lens distorsion, missing sections, inherent voxel-level dissimilarity between consecutive sections and the very large data-set size complicate the stitching procedure further.
The current registration strategy, together with a set of diagnostic tools that assist in monitoring the stitching process, is able to overcome all of the above difficulties and reconstruct cohesive ssTEM volumes at the scale of tens of terabytes for downstream processing, for example tracing and image segmentation. The procedure scales well on a compute cluster, and is currently employed at Janelia Research Campus – HHMI.
The tools consist of a core set of programs originally written in C/C++ by Bill Karsh and Lou Scheffer. Currently these tools are wrapped within an object oriented framework written in Matlab. The framework maximizes automation of the stitching prcedure, refines registration quality, manages job submission and job monitoring on the compute cluster, provides sanity checks at every step, and visual diagnostic tools.
Figure: A series of ssTEM sections through an adult fly brain aligned using the stitching tool. Each section is comprised of ~2000 individual tiles.
