The long-term goal of my group is to understand how neural representations of the sensory world generate learned behavioral responses. We are particularly interested in choice behaviors where sensory cues are evaluated based on multiple memories.
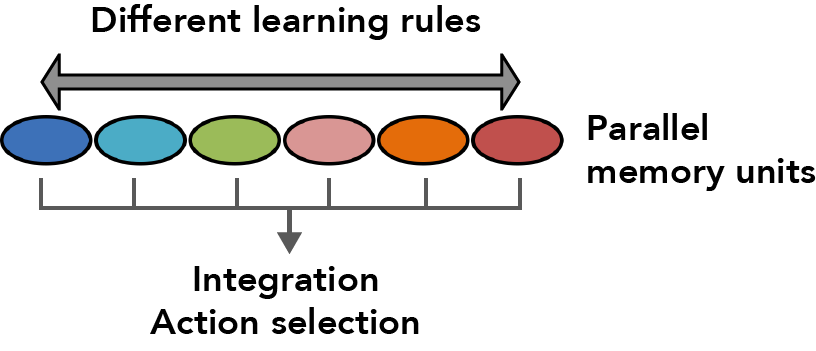
Animals learn the predictive value of stimuli based on temporal correlations with reward or punishment. This associative learning produces lasting changes in neural circuits referred to as memory traces or engrams. We know that multiple, parallel memory systems in different parts of the human brain (e.g. caudate nucleus, hippocampus, and amygdala) process and store different information during learning; similarly, in rodents, multiple, distributed circuits encode even simple memory forms such as fear conditioning. Do these multiple engrams serve different functions, and how are they integrated to control behavior? Fully localizing these distributed engrams, understanding what information is stored in each individual memory unit, and determining how units interact to function as one network are vital but highly challenging problems.
The mushroom body as a model for parallel memory units
Our study focuses on how circuits of the Drosophila mushroom body (MB) implement memory-based valuation of sensory cues and action selection. During associative olfactory learning, odor exposure (conditioned stimulus) in association with a reward or punishment (unconditioned stimulus) results in appetitive or aversive memory. Olfactory memory formation and retrieval in insects requires the MB. Available data strongly suggest that olfactory memories are stored as altered synaptic weights between Kenyon cells, the intrinsic neurons of the MB, and post-synaptic MB output neurons.
The first two layers of the fly olfactory system are highly analogous to vertebrates. Second order neurons relay olfactory information to the dendrites of approximately 2,000 MB Kenyon cells. The tiny Kenyon cell axons form parallel axon bundles that intersect with a small number of neurons with huge arbors, an arrangement strongly reminiscent of the mammalian cerebellum. Similarly, sparse odor coding across Kenyon cells maximizes the coding capacity of memories and minimizes interference between memories.
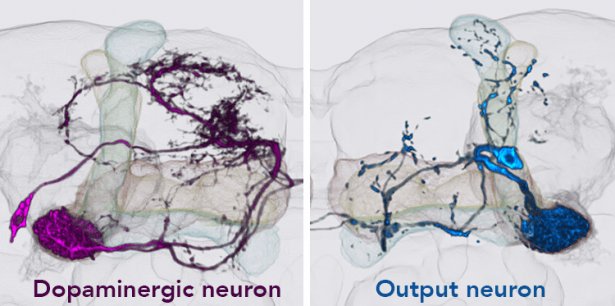
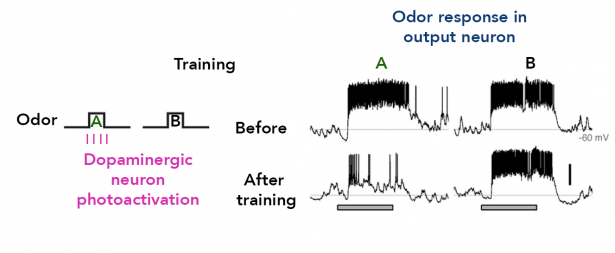
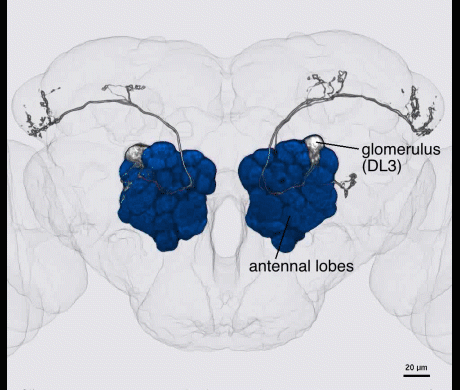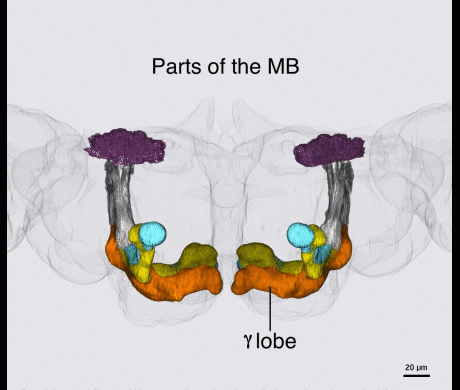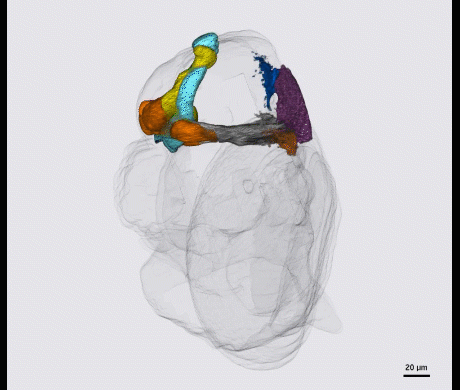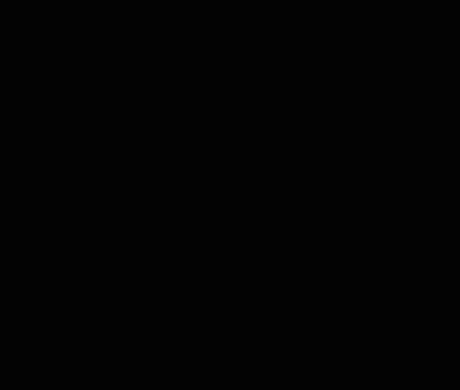 Kenyon cells are a site of convergence for olfactory information and reward or punishment signals delivered by dopaminergic neurons (DANs). 20 DAN types each innervate a compartmental region along the Kenyon cell parallel axonal fibers; these regions exactly match the dendritic arbors of 21 types of MB output neurons. Together DANs and MB output neurons therefore define anatomical compartments of the MB that are also units of associative learning. Punishment and reward signals are carried by distinct DANs targeting different compartments. DANs induce an enduring depression of just those Kenyon cell-output neuron synapses active in a compartment at the time of dopamine release. Thus, the compartment that receives dopamine during training determines memory valence, while the identity of Kenyon cells active during training determines sensory specificity.
Kenyon cells are a site of convergence for olfactory information and reward or punishment signals delivered by dopaminergic neurons (DANs). 20 DAN types each innervate a compartmental region along the Kenyon cell parallel axonal fibers; these regions exactly match the dendritic arbors of 21 types of MB output neurons. Together DANs and MB output neurons therefore define anatomical compartments of the MB that are also units of associative learning. Punishment and reward signals are carried by distinct DANs targeting different compartments. DANs induce an enduring depression of just those Kenyon cell-output neuron synapses active in a compartment at the time of dopamine release. Thus, the compartment that receives dopamine during training determines memory valence, while the identity of Kenyon cells active during training determines sensory specificity.
Using optogenetic activation of individual DAN cell types, we found extensive differences in the rate of memory formation, decay dynamics, storage capacity and flexibility to learn new associations across different MB compartments (Aso and Rubin, 2016). Thus, parallel memory units appear to implement different learning rules, providing multiple strategies to predict future events from the same past experiences.
Our lab seeks to understand the molecular and circuit basis of these different learning rules and how information from multiple memory units is integrated to guide action selection.
Ongoing Projects
Cotransmitters of DANs as a key regulator of learning rules
Our latest transcriptome data identified cell-type-specific cotransmitters of DANs. We found that some DANs can induce memories using cotransmitters in the absence of dopamine. The effect of cotransmitters is either synergistic or antagonistic to dopamine. Also, the dynamics of cotransmitter effects were clearly distinct from that of dopamine. Intriguingly, we could alter the learning rules of some MB compartments by swapping the cotransmitter. Our ongoing functional analysis of these cotransmitters will provide key mechanistic insights into the diversification of memory dynamics and learning rules in parallel memory systems.
Cellular biology of memory engrams in parallel memory units
What are the cellular bases of memory engrams? Despite the many genetic tools available for visualizing neurons and a long list of memory-related genes mapped onto Kenyon cells, the cellular biology of the Kenyon cells that store memories is largely unknown. The process of learning is thought to entail changes in the number of synapses as well as the compartment-specific changes in the distribution of synaptic proteins, channels, and receptors. However, it has been very difficult to study these cellular events because they are expected to occur only in the “memory engram cells” – the ~6% of Kenyon cells that are activated by a particular odor. Moreover, the fine axons of these cells are intermingled with other Kenyon cells that are not involved in the memory formation of that odor.
Now, this technical limitation is partly solved by the combined use of expansion and lattice light sheet microscopy.
In conjunction with CRISPR–tagging, intrabodies and fluorescence lifetime imaging microscopy (FLIM), we will elucidate biochemical/cellular events underlying distinct forms of synaptic plasticity in the MB.
Developing tools to inhibit/degrade proteins with subcellular specificity
EM connectome and transcriptome data can instantly provide many working hypotheses about the physiology of individual synapses and microcircuits. The collection of cell type-specific split–GAL4 driver lines allows for the manipulation of genes or neuronal activity. However, it is insufficient to precisely link EM–level structure to function.
We need to manipulate proteins at a subcellular level.
Circuit mechanisms integrating information from parallel memory units in behaving animals
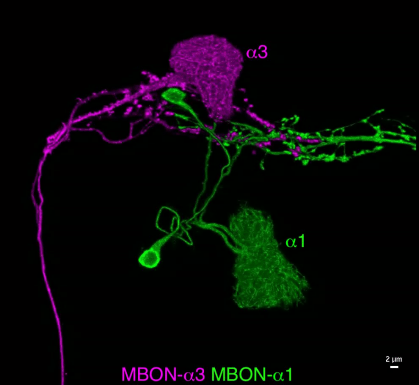
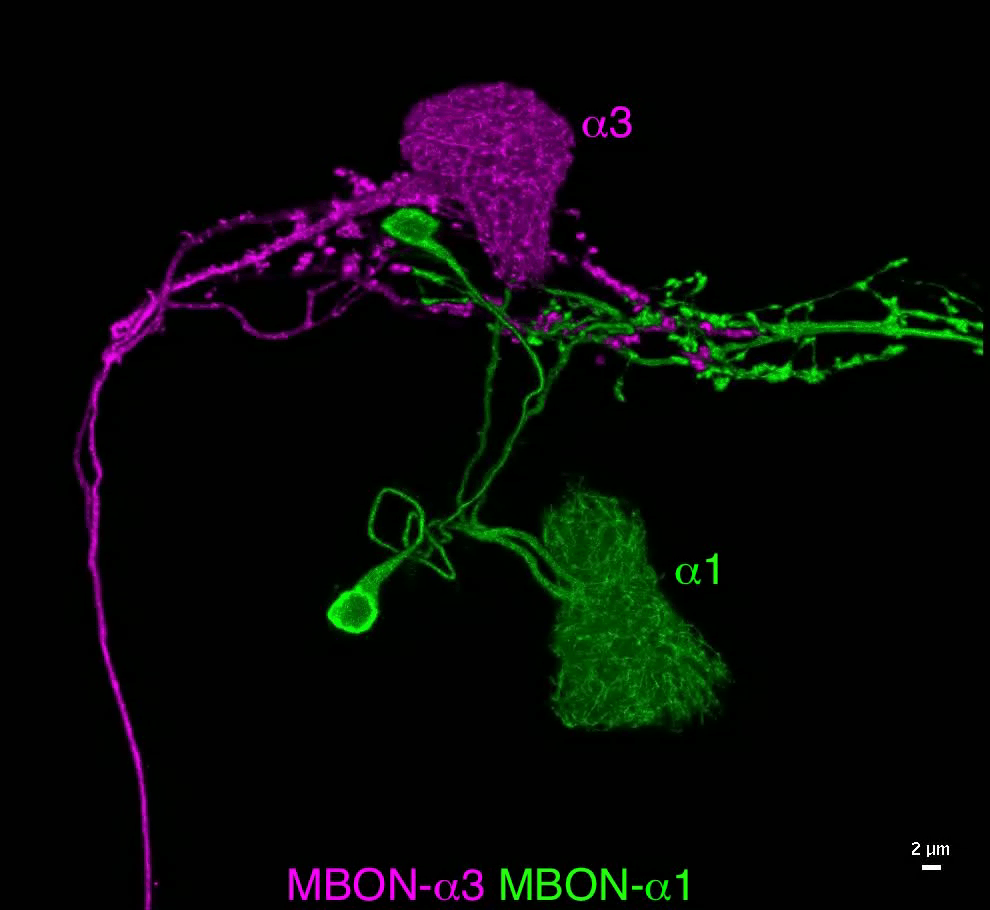
Parallel memory units in the MB store different information about a given odor. How are these distinct memories integrated to guide action selection? The axons of the output neurons converge in a brain region outside the MB. We anatomically searched for downstream neurons that receive converging inputs from MB output neurons, and then generated a set of genetic drivers to manipulate them. Now, we will fully utilize them to achieve our goal.
In certain situations, conflicting memories about the same sensory stimuli can be formed in parallel. For example, ethanol or foods containing nutritional sugar and bitterness can induce both appetitive and aversive memories (Das et al., 2014, Kaun et al., 2011). In such situations, the MB output neuron network seems to non-linearly integrate these conflicting signals for robust choice behaviors (Aso and Rubin, 2016). Even within the compartments of the same memory valence, distinct learning rules inevitably result in the storage of distinct information about the same experience. What is the circuit mechanism to integrate information from parallel memory units? How do the neurons downstream of the MB output neurons integrate signals from multiple MB output neurons? How reliably does their activity predict the future actions of flies? We will address these questions by combining neuroanatomy, behavioral assays and calcium imaging in behaving flies.
Computational model of distinct learning rules
We are collaborating with neurotheorists at the Center for Theoretical Neuroscience at Columbia University, including Ashok Litwin Kumar. We will systematically investigate how animals could benefit from how heterogeneous learning rules in parallel memory systems.