Filter
Associated Lab
- Aguilera Castrejon Lab (16) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (63) Apply Ahrens Lab filter
- Aso Lab (40) Apply Aso Lab filter
- Baker Lab (38) Apply Baker Lab filter
- Betzig Lab (112) Apply Betzig Lab filter
- Beyene Lab (13) Apply Beyene Lab filter
- Bock Lab (17) Apply Bock Lab filter
- Branson Lab (52) Apply Branson Lab filter
- Card Lab (42) Apply Card Lab filter
- Cardona Lab (63) Apply Cardona Lab filter
- Chklovskii Lab (13) Apply Chklovskii Lab filter
- Clapham Lab (14) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (12) Apply Darshan Lab filter
- Dennis Lab (1) Apply Dennis Lab filter
- Dickson Lab (46) Apply Dickson Lab filter
- Druckmann Lab (25) Apply Druckmann Lab filter
- Dudman Lab (50) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (11) Apply Egnor Lab filter
- Espinosa Medina Lab (19) Apply Espinosa Medina Lab filter
- Feliciano Lab (7) Apply Feliciano Lab filter
- Fetter Lab (41) Apply Fetter Lab filter
- Fitzgerald Lab (29) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (38) Apply Funke Lab filter
- Gonen Lab (91) Apply Gonen Lab filter
- Grigorieff Lab (62) Apply Grigorieff Lab filter
- Harris Lab (60) Apply Harris Lab filter
- Heberlein Lab (94) Apply Heberlein Lab filter
- Hermundstad Lab (26) Apply Hermundstad Lab filter
- Hess Lab (77) Apply Hess Lab filter
- Ilanges Lab (2) Apply Ilanges Lab filter
- Jayaraman Lab (46) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (6) Apply Johnson Lab filter
- Kainmueller Lab (19) Apply Kainmueller Lab filter
- Karpova Lab (14) Apply Karpova Lab filter
- Keleman Lab (13) Apply Keleman Lab filter
- Keller Lab (76) Apply Keller Lab filter
- Koay Lab (18) Apply Koay Lab filter
- Lavis Lab (148) Apply Lavis Lab filter
- Lee (Albert) Lab (34) Apply Lee (Albert) Lab filter
- Leonardo Lab (23) Apply Leonardo Lab filter
- Li Lab (28) Apply Li Lab filter
- Lippincott-Schwartz Lab (168) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (6) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (61) Apply Liu (Zhe) Lab filter
- Looger Lab (138) Apply Looger Lab filter
- Magee Lab (49) Apply Magee Lab filter
- Menon Lab (18) Apply Menon Lab filter
- Murphy Lab (13) Apply Murphy Lab filter
- O'Shea Lab (6) Apply O'Shea Lab filter
- Otopalik Lab (13) Apply Otopalik Lab filter
- Pachitariu Lab (47) Apply Pachitariu Lab filter
- Pastalkova Lab (18) Apply Pastalkova Lab filter
- Pavlopoulos Lab (19) Apply Pavlopoulos Lab filter
- Pedram Lab (15) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (51) Apply Reiser Lab filter
- Riddiford Lab (44) Apply Riddiford Lab filter
- Romani Lab (43) Apply Romani Lab filter
- Rubin Lab (143) Apply Rubin Lab filter
- Saalfeld Lab (63) Apply Saalfeld Lab filter
- Satou Lab (16) Apply Satou Lab filter
- Scheffer Lab (36) Apply Scheffer Lab filter
- Schreiter Lab (67) Apply Schreiter Lab filter
- Sgro Lab (21) Apply Sgro Lab filter
- Shroff Lab (31) Apply Shroff Lab filter
- Simpson Lab (23) Apply Simpson Lab filter
- Singer Lab (80) Apply Singer Lab filter
- Spruston Lab (93) Apply Spruston Lab filter
- Stern Lab (156) Apply Stern Lab filter
- Sternson Lab (54) Apply Sternson Lab filter
- Stringer Lab (35) Apply Stringer Lab filter
- Svoboda Lab (135) Apply Svoboda Lab filter
- Tebo Lab (33) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (21) Apply Tillberg Lab filter
- Tjian Lab (64) Apply Tjian Lab filter
- Truman Lab (88) Apply Truman Lab filter
- Turaga Lab (51) Apply Turaga Lab filter
- Turner Lab (37) Apply Turner Lab filter
- Vale Lab (7) Apply Vale Lab filter
- Voigts Lab (3) Apply Voigts Lab filter
- Wang (Meng) Lab (19) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (25) Apply Wang (Shaohe) Lab filter
- Wu Lab (9) Apply Wu Lab filter
- Zlatic Lab (28) Apply Zlatic Lab filter
- Zuker Lab (25) Apply Zuker Lab filter
Associated Project Team
- CellMap (12) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- FIB-SEM Technology (2) Apply FIB-SEM Technology filter
- Fly Descending Interneuron (11) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (53) Apply FlyEM filter
- FlyLight (49) Apply FlyLight filter
- GENIE (45) Apply GENIE filter
- Integrative Imaging (3) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (18) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (26) Apply Tool Translation Team (T3) filter
- Transcription Imaging (49) Apply Transcription Imaging filter
Publication Date
- 2025 (102) Apply 2025 filter
- 2024 (219) Apply 2024 filter
- 2023 (160) Apply 2023 filter
- 2022 (193) Apply 2022 filter
- 2021 (194) Apply 2021 filter
- 2020 (196) Apply 2020 filter
- 2019 (202) Apply 2019 filter
- 2018 (232) Apply 2018 filter
- 2017 (217) Apply 2017 filter
- 2016 (209) Apply 2016 filter
- 2015 (252) Apply 2015 filter
- 2014 (236) Apply 2014 filter
- 2013 (194) Apply 2013 filter
- 2012 (190) Apply 2012 filter
- 2011 (190) Apply 2011 filter
- 2010 (161) Apply 2010 filter
- 2009 (158) Apply 2009 filter
- 2008 (140) Apply 2008 filter
- 2007 (106) Apply 2007 filter
- 2006 (92) Apply 2006 filter
- 2005 (67) Apply 2005 filter
- 2004 (57) Apply 2004 filter
- 2003 (58) Apply 2003 filter
- 2002 (39) Apply 2002 filter
- 2001 (28) Apply 2001 filter
- 2000 (29) Apply 2000 filter
- 1999 (14) Apply 1999 filter
- 1998 (18) Apply 1998 filter
- 1997 (16) Apply 1997 filter
- 1996 (10) Apply 1996 filter
- 1995 (18) Apply 1995 filter
- 1994 (12) Apply 1994 filter
- 1993 (10) Apply 1993 filter
- 1992 (6) Apply 1992 filter
- 1991 (11) Apply 1991 filter
- 1990 (11) Apply 1990 filter
- 1989 (6) Apply 1989 filter
- 1988 (1) Apply 1988 filter
- 1987 (7) Apply 1987 filter
- 1986 (4) Apply 1986 filter
- 1985 (5) Apply 1985 filter
- 1984 (2) Apply 1984 filter
- 1983 (2) Apply 1983 filter
- 1982 (3) Apply 1982 filter
- 1981 (3) Apply 1981 filter
- 1980 (1) Apply 1980 filter
- 1979 (1) Apply 1979 filter
- 1976 (2) Apply 1976 filter
- 1973 (1) Apply 1973 filter
- 1970 (1) Apply 1970 filter
- 1967 (1) Apply 1967 filter
Type of Publication
4087 Publications
Showing 2361-2370 of 4087 resultsMeiosis is a highly conserved process in which a diploid genome is recombined and assorted into haploid gametes. Remarkably, the pea aphid Acyrthosiphon pisum exhibits a reproductive polyphenism whereby environmental signals trigger a switch between apomixis (parthenogenetic reproduction) and meiosis (sexual reproduction). Aphid apomixis results in daughter embryo clones with 2n genome content without male contribution or recombination. This important adaptation allows aphid populations to not only rapidly expand upon abundant resources during summer but also survive winter. How aphids have evolved this ability to switch between parthenogenesis and sexual meiosis is unknown. To arrive at a mechanistic explanation for this developmental plasticity, I determined meiosis gene activity in sexuals and asexuals. I first identified homologs of a core set of meiosis genes from the pea aphid genome. Next, I tested the expression of these core meiosis genes by PCR spanning across at least one intron from cDNA isolated from asexual and sexual ovaries. Surprisingly, meiosis specific genes (e.g., Spo11, Msh4, Msh5, Hop2 and Mnd1) are expressed in asexual ovaries. Additionally, the Spo11 PCR product contained intronic sequence, thus representing unspliced mRNA. Future experiments looking at the quantities and localizations of mRNA and protein will help to distinguish among several possible explanations for these results. Further molecular characterization of this phenotypic plasticity will be helpful in understanding how multiple interacting pathways can evolve to create alternate developmental phenotypes.
Projection neurons (PNs) in the mammalian olfactory bulb (OB) receive input from the nose and project to diverse cortical and subcortical areas. Morphological and physiological studies have highlighted functional heterogeneity, yet no molecular markers have been described that delineate PN subtypes. Here, we used viral injections into olfactory cortex and fluorescent nucleus sorting to enrich PNs for high-throughput single nucleus and bulk RNA deep sequencing. Transcriptome analysis and RNA hybridization identified distinct mitral and tufted cell populations with characteristic transcription factor network topology, cell adhesion and excitability-related gene expression. Finally, we describe a new computational approach for integrating bulk and snRNA-seq data, and provide evidence that different mitral cell populations preferentially project to different target regions. Together, we have identified potential molecular and gene regulatory mechanisms underlying PN diversity and provide new molecular entry points into studying the diverse functional roles of mitral and tufted cell subtypes.
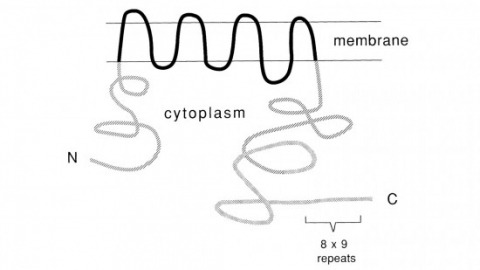
Recent studies suggest that the fly uses the inositol lipid signaling system for visual excitation and that the Drosophila transient receptor potential (trp) mutation disrupts this process subsequent to the production of IP3. In this paper, we show that trp encodes a novel 1275 amino acid protein with eight putative transmembrane segments. Immunolocalization indicates that the trp protein is expressed predominantly in the rhabdomeric membranes of the photoreceptor cells.
Most traditional optical biosensors operate through molecular recognition, where ligand binding causes conformational changes that lead to optical perturbations in the emitting motif. Optical sensors developed from single-stranded DNA-functionalized single-walled carbon nanotubes (ssDNA–SWCNTs) have started to make useful contributions to biological research. However, the mechanisms underlying their function have remained poorly understood. In this study, we combine experimental and computational approaches to show that ligand binding alone is not sufficient for optical modulation in this class of synthetic biosensors. Instead, the optical response that occurs after ligand binding is highly dependent on the chemical properties of the ligands, resembling mechanisms seen in activity-based biosensors. Specifically, we show that in ssDNA–SWCNT catecholamine sensors, the optical response correlates positively with the electron density on the aryl motif, even among ligands with similar ligand binding affinities. Importantly, despite the strong correlations with electrochemical properties, we find that catechol oxidation itself is not necessary to drive the sensor optical response. We discuss how these findings could serve as a framework for tuning the performance of existing sensors and guiding the development of new biosensors of this class.
Cell recognition requires interactions through molecules located on cell surface. The insect homolog of Down syndrome cell adhesion molecule (Dscam) manifests huge molecular diversity in its extracellular domain. High-affinity Dscam-Dscam interactions only occur between isoforms that carry identical extracellular domains. Homophilic Dscam signaling can, thus, vary in strength depending on the compositions of Dscams present on the opposing cell surfaces. Dscam abundantly exists in the developing nervous system and governs arborization and proper elaboration of neurites. Notably, individual neurons may stochastically and dynamically express a small subset of Dscam isoforms such that any given neurite can be endowed with a unique repertoire of Dscams. This allows individual neurites to recognize their sister branches. Self-recognition leads to self-repulsion, ensuring divergent migration of sister processes. By contrast, weak homophilic Dscam interactions may promote fasciculation of neurites that express analogous, but not identical, Dscams. Differential Dscam binding may provide graded cell recognition that in turn governs complex neuronal morphogenesis.
Glucose transporters (GLUTs) provide a pathway for glucose transport across membranes. Human GLUTs are implicated in devastating diseases such as heart disease, hyper- and hypo-glycemia, type 2 diabetes and caner. The human GLUT1 has been recently crystalized in the inward-facing open conformation. However, there is no other structural information for other conformations. The X-ray structures of E. coli Xylose permease (XylE), a glucose transporter homolog, are available in multiple conformations with and without the substrates D-xylose and D-glucose. XylE has high sequence homology to human GLUT1 and key residues in the sugar-binding pocket are conserved. Here we construct a homology model for human GLUT1 based on the available XylE crystal structure in the partially occluded outward-facing conformation. A long unbiased all atom molecular dynamics simulation starting from the model can capture a new fully opened outward-facing conformation. Our investigation of molecular interactions at the interface between the transmembrane (TM) domains and the intracellular helices (ICH) domain in the outward- and inward-facing conformation supports that the ICH domain likely stabilizes the outward-facing conformation in GLUT1. Furthermore, inducing a conformational transition, our simulations manifest a global asymmetric rocker switch motion and detailed molecular interactions between the substrate and residues through the water-filled selective pore along a pathway from the extracellular to the intracellular side. The results presented here are consistent with previously published biochemical, mutagenesis and functional studies. Together, this study shed light on the structure and functional relationships of GLUT1 in multiple conformational states.
Recently, the fruit fly Drosophila melanogaster has been introduced as a model system to study the molecular bases of a variety of ethanol-induced behaviors. It became immediately apparent that the behavioral changes elicited by acute ethanol exposure are remarkably similar in flies and mammals. Flies show signs of acute intoxication, which range from locomotor stimulation at low doses to complete sedation at higher doses and they develop tolerance upon intermittent ethanol exposure. Genetic screens for mutants with altered responsiveness to ethanol have been carried out and a few of the disrupted genes have been identified. This analysis, while still in its early stages, has already revealed some surprising molecular parallels with mammals. The availability of powerful tools for genetic manipulation in Drosophila, together with the high degree of conservation at the genomic level, make Drosophila a promising model organism to study the mechanism by which ethanol regulates behavior and the mechanisms underlying the organism's adaptation to long-term ethanol exposure.
In Saccharomyces cerevisiae, the repressor Crt1 and the global corepressor Ssn6-Tup1 repress the DNA damage-inducible ribonucleotide reductase (RNR) genes. Initiation of DNA damage signals causes the release of Crt1 and Ssn6-Tup1 from the promoter, coactivator recruitment, and derepression of transcription, indicating that Crt1 plays a crucial role in the switch between gene repression and activation. Here we have mapped the functional domains of Crt1 and identified two independent repression domains and a region required for gene activation. The N terminus of Crt1 is the major repression domain, it directly binds to the Ssn6-Tup1 complex, and its repression activities are dependent upon Ssn6-Tup1 and histone deacetylases (HDACs). In addition, we identified a C-terminal repression domain, which is independent of Ssn6-Tup1 and HDACs and functions at native genes in vivo. Furthermore, we show that TFIID and SWI/SNF bind to a region within the N terminus of Crt1, overlapping with but distinct from the Ssn6-Tup1 binding and repression domain, suggesting that Crt1 may have activator functions. Crt1 mutants were constructed to dissect its activator and repressor functions. All of the mutants were competent for repression of the DNA damage-inducible genes, but a majority were "derepression-defective" mutants. Further characterization of these mutants indicated that they are capable of receiving DNA damage signals and releasing the Ssn6-Tup1 complex from the promoter but are selectively impaired for TFIID and SWI/SNF recruitment. These results imply a two-step activation model of the DNA damage-inducible genes and that Crt1 functions as a signal-dependent dual-transcription activator and repressor that acts in a transient manner.
Rotaviruses, major causes of childhood gastroenteritis, are nonenveloped, icosahedral particles with double-strand RNA genomes. By the use of electron cryomicroscopy and single-particle reconstruction, we have visualized a rotavirus particle comprising the inner capsid coated with the trimeric outer-layer protein, VP7, at a resolution (4 A) comparable with that of X-ray crystallography. We have traced the VP7 polypeptide chain, including parts not seen in its X-ray crystal structure. The 3 well-ordered, 30-residue, N-terminal "arms" of each VP7 trimer grip the underlying trimer of VP6, an inner-capsid protein. Structural differences between free and particle-bound VP7 and between free and VP7-coated inner capsids may regulate mRNA transcription and release. The Ca(2+)-stabilized VP7 intratrimer contact region, which presents important neutralizing epitopes, is unaltered upon capsid binding.