Filter
Associated Lab
- Aguilera Castrejon Lab (1) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (4) Apply Ahrens Lab filter
- Aso Lab (3) Apply Aso Lab filter
- Betzig Lab (4) Apply Betzig Lab filter
- Beyene Lab (1) Apply Beyene Lab filter
- Branson Lab (3) Apply Branson Lab filter
- Card Lab (5) Apply Card Lab filter
- Cardona Lab (3) Apply Cardona Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Dickson Lab (4) Apply Dickson Lab filter
- Dudman Lab (2) Apply Dudman Lab filter
- Espinosa Medina Lab (2) Apply Espinosa Medina Lab filter
- Fitzgerald Lab (3) Apply Fitzgerald Lab filter
- Funke Lab (3) Apply Funke Lab filter
- Grigorieff Lab (3) Apply Grigorieff Lab filter
- Harris Lab (1) Apply Harris Lab filter
- Heberlein Lab (2) Apply Heberlein Lab filter
- Hermundstad Lab (2) Apply Hermundstad Lab filter
- Hess Lab (5) Apply Hess Lab filter
- Jayaraman Lab (4) Apply Jayaraman Lab filter
- Johnson Lab (1) Apply Johnson Lab filter
- Keller Lab (5) Apply Keller Lab filter
- Lavis Lab (9) Apply Lavis Lab filter
- Lee (Albert) Lab (5) Apply Lee (Albert) Lab filter
- Li Lab (3) Apply Li Lab filter
- Lippincott-Schwartz Lab (8) Apply Lippincott-Schwartz Lab filter
- Liu (Zhe) Lab (7) Apply Liu (Zhe) Lab filter
- Looger Lab (7) Apply Looger Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Pachitariu Lab (2) Apply Pachitariu Lab filter
- Pedram Lab (3) Apply Pedram Lab filter
- Podgorski Lab (5) Apply Podgorski Lab filter
- Reiser Lab (2) Apply Reiser Lab filter
- Romani Lab (2) Apply Romani Lab filter
- Rubin Lab (9) Apply Rubin Lab filter
- Saalfeld Lab (2) Apply Saalfeld Lab filter
- Satou Lab (1) Apply Satou Lab filter
- Scheffer Lab (1) Apply Scheffer Lab filter
- Schreiter Lab (5) Apply Schreiter Lab filter
- Sgro Lab (4) Apply Sgro Lab filter
- Spruston Lab (5) Apply Spruston Lab filter
- Stern Lab (4) Apply Stern Lab filter
- Sternson Lab (4) Apply Sternson Lab filter
- Stringer Lab (2) Apply Stringer Lab filter
- Svoboda Lab (5) Apply Svoboda Lab filter
- Tebo Lab (4) Apply Tebo Lab filter
- Truman Lab (3) Apply Truman Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
- Turner Lab (3) Apply Turner Lab filter
- Wang (Shaohe) Lab (1) Apply Wang (Shaohe) Lab filter
- Zlatic Lab (2) Apply Zlatic Lab filter
Associated Project Team
- Fly Descending Interneuron (2) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (1) Apply Fly Functional Connectome filter
- FlyEM (3) Apply FlyEM filter
- FlyLight (8) Apply FlyLight filter
- GENIE (5) Apply GENIE filter
- MouseLight (1) Apply MouseLight filter
- Tool Translation Team (T3) (3) Apply Tool Translation Team (T3) filter
- Transcription Imaging (1) Apply Transcription Imaging filter
Publication Date
- Remove 2020 filter 2020
Type of Publication
196 Publications
Showing 41-50 of 196 resultsBrains encode behaviors using neurons amenable to systematic classification by gene expression. The contribution of molecular identity to neural coding is not understood because of the challenges involved with measuring neural dynamics and molecular information from the same cells. We developed CaRMA (calcium and RNA multiplexed activity) imaging based on recording in vivo single-neuron calcium dynamics followed by gene expression analysis. We simultaneously monitored activity in hundreds of neurons in mouse paraventricular hypothalamus (PVH). Combinations of cell-type marker genes had predictive power for neuronal responses across 11 behavioral states. The PVH uses combinatorial assemblies of molecularly defined neuron populations for grouped-ensemble coding of survival behaviors. The neuropeptide receptor neuropeptide Y receptor type 1 (Npy1r) amalgamated multiple cell types with similar responses. Our results show that molecularly defined neurons are important processing units for brain function.
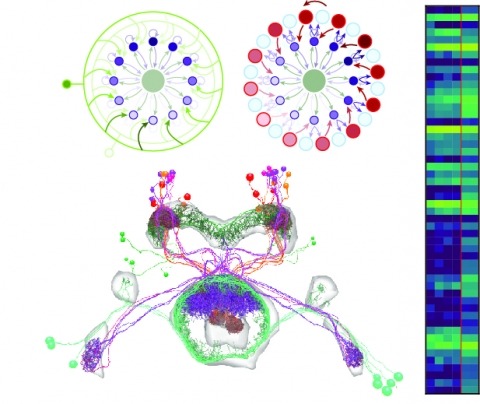
Neural representations of head direction (HD) have been discovered in many species. Theoretical work has proposed that the dynamics associated with these representations are generated, maintained, and updated by recurrent network structures called ring attractors. We evaluated this theorized structure-function relationship by performing electron-microscopy-based circuit reconstruction and RNA profiling of identified cell types in the HD system of Drosophila melanogaster. We identified motifs that have been hypothesized to maintain the HD representation in darkness, update it when the animal turns, and tether it to visual cues. Functional studies provided support for the proposed roles of individual excitatory or inhibitory circuit elements in shaping activity. We also discovered recurrent connections between neuronal arbors with mixed pre- and postsynaptic specializations. Our results confirm that the Drosophila HD network contains the core components of a ring attractor while also revealing unpredicted structural features that might enhance the network's computational power.
Fluorescent biosensors are powerful tools for the detection of biochemical events inside cells with high spatiotemporal resolution. Biosensors based on fluorescent proteins often suffer from issues with photostability and brightness. On the other hand, hybrid, chemical–genetic systems present unique opportunities to combine the strengths of synthetic, organic chemistry with biological macromolecules to generate exquisitely tailored semisynthetic sensors.
An important question in early neural development is the origin of stochastic nuclear movement between apical and basal surfaces of neuroepithelia during interkinetic nuclear migration. Tracking of nuclear subpopulations has shown evidence of diffusion - mean squared displacements growing linearly in time - and suggested crowding from cell division at the apical surface drives basalward motion. Yet, this hypothesis has not yet been tested, and the forces involved not quantified. We employ long-term, rapid light-sheet and two-photon imaging of early zebrafish retinogenesis to track entire populations of nuclei within the tissue. The time-varying concentration profiles show clear evidence of crowding as nuclei reach close-packing and are quantitatively described by a nonlinear diffusion model. Considerations of nuclear motion constrained inside the enveloping cell membrane show that concentration-dependent stochastic forces inside cells, compatible in magnitude to those found in cytoskeletal transport, can explain the observed magnitude of the diffusion constant.
mTOR is a serine/threonine kinase and a master regulator of cell growth and proliferation. Raptor, a scaffolding protein that recruits substrates to mTOR complex 1 (mTORC1), is known to be phosphorylated during mitosis, but the significance of this phosphorylation remains largely unknown. Here we show that raptor expression and mTORC1 activity are dramatically reduced in cells arrested in mitosis. Expression of a non-phosphorylatable raptor mutant reactivates mTORC1 and significantly reduces cytotoxicity of the mitotic poison Taxol. This effect is mediated via degradation of PDCD4, a tumor suppressor protein that inhibits eIF4A activity and is negatively regulated by the mTORC1/S6K pathway. Moreover, pharmacological inhibition of eIF4A is able to enhance the effects of Taxol and restore sensitivity in Taxol-resistant cancer cells. These findings indicate that the mTORC1/S6K/PDCD4/eIF4A axis has a pivotal role in the death versus slippage decision during mitotic arrest and may be exploited clinically to treat tumors resistant to anti-mitotic agents.
The mating decisions of Drosophila melanogaster females are primarily revealed through either of two discrete actions: opening of the vaginal plates to allow copulation, or extrusion of the ovipositor to reject the male. Both actions are triggered by the male courtship song, and both are dependent upon the female's mating status. Virgin females are more likely to open their vaginal plates in response to song; mated females are more likely to extrude their ovipositor. Here, we examine the neural cause and behavioral consequence of ovipositor extrusion. We show that the DNp13 descending neurons act as command-type neurons for ovipositor extrusion, and that ovipositor extrusion is an effective deterrent only when performed by females that have previously mated. The DNp13 neurons respond to male song via direct synaptic input from the pC2l auditory neurons. Mating status does not modulate the song responses of DNp13 neurons, but rather how effectively they can engage the motor circuits for ovipositor extrusion. We present evidence that mating status information is mediated by ppk sensory neurons in the uterus, which are activated upon ovulation. Vaginal plate opening and ovipositor extrusion are thus controlled by anatomically and functionally distinct circuits, highlighting the diversity of neural decision-making circuits even in the context of closely related behaviors with shared exteroceptive and interoceptive inputs.
The striosome compartment within the dorsal striatum has been implicated in reinforcement learning and regulation of motivation, but how striosomal neurons contribute to these functions remains elusive. Here, we show that a genetically identified striosomal population, which expresses the Teashirt family zinc finger 1 (Tshz1) and belongs to the direct pathway, drives negative reinforcement and is essential for aversive learning in mice. Contrasting a "conventional" striosomal direct pathway, the Tshz1 neurons cause aversion, movement suppression, and negative reinforcement once activated, and they receive a distinct set of synaptic inputs. These neurons are predominantly excited by punishment rather than reward and represent the anticipation of punishment or the motivation for avoidance. Furthermore, inhibiting these neurons impairs punishment-based learning without affecting reward learning or movement. These results establish a major role of striosomal neurons in behaviors reinforced by punishment and moreover uncover functions of the direct pathway unaccounted for in classic models.