Filter
Associated Lab
- Aso Lab (30) Apply Aso Lab filter
- Betzig Lab (1) Apply Betzig Lab filter
- Bock Lab (2) Apply Bock Lab filter
- Branson Lab (8) Apply Branson Lab filter
- Card Lab (5) Apply Card Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Dickson Lab (2) Apply Dickson Lab filter
- Druckmann Lab (1) Apply Druckmann Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Funke Lab (1) Apply Funke Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (2) Apply Heberlein Lab filter
- Hermundstad Lab (2) Apply Hermundstad Lab filter
- Hess Lab (5) Apply Hess Lab filter
- Jayaraman Lab (6) Apply Jayaraman Lab filter
- Lippincott-Schwartz Lab (1) Apply Lippincott-Schwartz Lab filter
- Looger Lab (2) Apply Looger Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Reiser Lab (15) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Romani Lab (1) Apply Romani Lab filter
- Remove Rubin Lab filter Rubin Lab
- Saalfeld Lab (4) Apply Saalfeld Lab filter
- Scheffer Lab (7) Apply Scheffer Lab filter
- Schreiter Lab (1) Apply Schreiter Lab filter
- Simpson Lab (3) Apply Simpson Lab filter
- Singer Lab (1) Apply Singer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Stern Lab (1) Apply Stern Lab filter
- Svoboda Lab (3) Apply Svoboda Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Truman Lab (4) Apply Truman Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
- Turner Lab (5) Apply Turner Lab filter
- Zuker Lab (1) Apply Zuker Lab filter
Associated Project Team
- CellMap (1) Apply CellMap filter
- Fly Functional Connectome (4) Apply Fly Functional Connectome filter
- Fly Olympiad (3) Apply Fly Olympiad filter
- FlyEM (11) Apply FlyEM filter
- FlyLight (20) Apply FlyLight filter
- GENIE (1) Apply GENIE filter
- Transcription Imaging (1) Apply Transcription Imaging filter
Publication Date
- 2025 (4) Apply 2025 filter
- 2024 (4) Apply 2024 filter
- 2023 (5) Apply 2023 filter
- 2022 (1) Apply 2022 filter
- 2021 (4) Apply 2021 filter
- 2020 (9) Apply 2020 filter
- 2019 (6) Apply 2019 filter
- 2018 (7) Apply 2018 filter
- 2017 (15) Apply 2017 filter
- 2016 (3) Apply 2016 filter
- 2015 (16) Apply 2015 filter
- 2014 (9) Apply 2014 filter
- 2013 (5) Apply 2013 filter
- 2012 (8) Apply 2012 filter
- 2011 (4) Apply 2011 filter
- 2010 (4) Apply 2010 filter
- 2009 (2) Apply 2009 filter
- 2008 (4) Apply 2008 filter
- 2007 (2) Apply 2007 filter
- 2006 (1) Apply 2006 filter
- 2002 (1) Apply 2002 filter
- 2000 (2) Apply 2000 filter
- 1999 (1) Apply 1999 filter
- 1997 (1) Apply 1997 filter
- 1995 (2) Apply 1995 filter
- 1994 (2) Apply 1994 filter
- 1993 (2) Apply 1993 filter
- 1992 (1) Apply 1992 filter
- 1991 (2) Apply 1991 filter
- 1990 (3) Apply 1990 filter
- 1989 (2) Apply 1989 filter
- 1987 (2) Apply 1987 filter
- 1986 (1) Apply 1986 filter
- 1985 (1) Apply 1985 filter
- 1984 (1) Apply 1984 filter
- 1983 (1) Apply 1983 filter
- 1982 (2) Apply 1982 filter
- 1981 (1) Apply 1981 filter
- 1979 (1) Apply 1979 filter
- 1973 (1) Apply 1973 filter
Type of Publication
143 Publications
Showing 71-80 of 143 resultsWe describe an engineered family of highly antigenic molecules based on GFP-like fluorescent proteins. These molecules contain numerous copies of peptide epitopes and simultaneously bind IgG antibodies at each location. These 'spaghetti monster' fluorescent proteins (smFPs) distributed well in neurons, notably into small dendrites, spines and axons. smFP immunolabeling localized weakly expressed proteins not well resolved with traditional epitope tags. By varying epitope and scaffold, we generated a diverse family of mutually orthogonal antigens. In cultured neurons and mouse and fly brains, smFP probes allowed robust, orthogonal multicolor visualization of proteins, cell populations and neuropil. smFP variants complement existing tracers and greatly increase the number of simultaneous imaging channels, and they performed well in advanced preparations such as array tomography, super-resolution fluorescence imaging and electron microscopy. In living cells, the probes improved single-molecule image tracking and increased yield for RNA-seq. These probes facilitate new experiments in connectomics, transcriptomics and protein localization.
Ends-out gene targeting allows seamless replacement of endogenous genes with engineered DNA fragments by homologous recombination, thus creating designer "genes" in the endogenous locus. Conventional gene targeting in Drosophila involves targeting with the preintegrated donor DNA in the larval primordial germ cells. Here we report G: ene targeting during O: ogenesis with L: ethality I: nhibitor and C: RISPR/Cas (Golic+), which improves on all major steps in such transgene-based gene targeting systems. First, donor DNA is integrated into precharacterized attP sites for efficient flip-out. Second, FLP, I-SceI, and Cas9 are specifically expressed in cystoblasts, which arise continuously from female germline stem cells, thereby providing a continual source of independent targeting events in each offspring. Third, a repressor-based lethality selection is implemented to facilitate screening for correct targeting events. Altogether, Golic+ realizes high-efficiency ends-out gene targeting in ovarian cystoblasts, which can be readily scaled up to achieve high-throughput genome editing.
Drosophila melanogaster plays an important role in molecular, genetic, and genomic studies of heredity, development, metabolism, behavior, and human disease. The initial reference genome sequence reported more than a decade ago had a profound impact on progress in Drosophila research, and improving the accuracy and completeness of this sequence continues to be important to further progress. We previously described improvement of the 117-Mb sequence in the euchromatic portion of the genome and 21 Mb in the heterochromatic portion, using a whole-genome shotgun assembly, BAC physical mapping, and clone-based finishing. Here, we report an improved reference sequence of the single-copy and middle-repetitive regions of the genome, produced using cytogenetic mapping to mitotic and polytene chromosomes, clone-based finishing and BAC fingerprint verification, ordering of scaffolds by alignment to cDNA sequences, incorporation of other map and sequence data, and validation by whole-genome optical restriction mapping. These data substantially improve the accuracy and completeness of the reference sequence and the order and orientation of sequence scaffolds into chromosome arm assemblies. Representation of the Y chromosome and other heterochromatic regions is particularly improved. The new 143.9-Mb reference sequence, designated Release 6, effectively exhausts clone-based technologies for mapping and sequencing. Highly repeat-rich regions, including large satellite blocks and functional elements such as the ribosomal RNA genes and the centromeres, are largely inaccessible to current sequencing and assembly methods and remain poorly represented. Further significant improvements will require sequencing technologies that do not depend on molecular cloning and that produce very long reads.
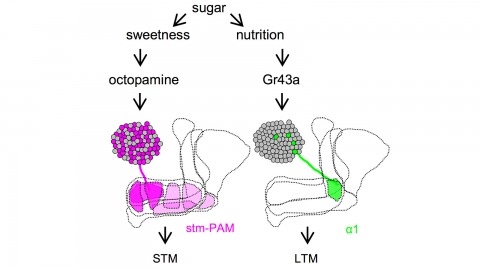
Drosophila melanogaster can acquire a stable appetitive olfactory memory when the presentation of a sugar reward and an odor are paired. However, the neuronal mechanisms by which a single training induces long-term memory are poorly understood. Here we show that two distinct subsets of dopamine neurons in the fly brain signal reward for short-term (STM) and long-term memories (LTM). One subset induces memory that decays within several hours, whereas the other induces memory that gradually develops after training. They convey reward signals to spatially segregated synaptic domains of the mushroom body (MB), a potential site for convergence. Furthermore, we identified a single type of dopamine neuron that conveys the reward signal to restricted subdomains of the mushroom body lobes and induces long-term memory. Constant appetitive memory retention after a single training session thus comprises two memory components triggered by distinct dopamine neurons.
Animals discriminate stimuli, learn their predictive value and use this knowledge to modify their behavior. In Drosophila, the mushroom body (MB) plays a key role in these processes. Sensory stimuli are sparsely represented by ∼2000 Kenyon cells, which converge onto 34 output neurons (MBONs) of 21 types. We studied the role of MBONs in several associative learning tasks and in sleep regulation, revealing the extent to which information flow is segregated into distinct channels and suggesting possible roles for the multi-layered MBON network. We also show that optogenetic activation of MBONs can, depending on cell type, induce repulsion or attraction in flies. The behavioral effects of MBON perturbation are combinatorial, suggesting that the MBON ensemble collectively represents valence. We propose that local, stimulus-specific dopaminergic modulation selectively alters the balance within the MBON network for those stimuli. Our results suggest that valence encoded by the MBON ensemble biases memory-based action selection.
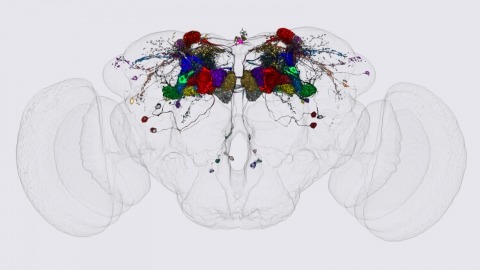
We identified the neurons comprising the Drosophila mushroom body (MB), an associative center in invertebrate brains, and provide a comprehensive map describing their potential connections. Each of the 21 MB output neuron (MBON) types elaborates segregated dendritic arbors along the parallel axons of ∼2000 Kenyon cells, forming 15 compartments that collectively tile the MB lobes. MBON axons project to five discrete neuropils outside of the MB and three MBON types form a feedforward network in the lobes. Each of the 20 dopaminergic neuron (DAN) types projects axons to one, or at most two, of the MBON compartments. Convergence of DAN axons on compartmentalized Kenyon cell-MBON synapses creates a highly ordered unit that can support learning to impose valence on sensory representations. The elucidation of the complement of neurons of the MB provides a comprehensive anatomical substrate from which one can infer a functional logic of associative olfactory learning and memory.
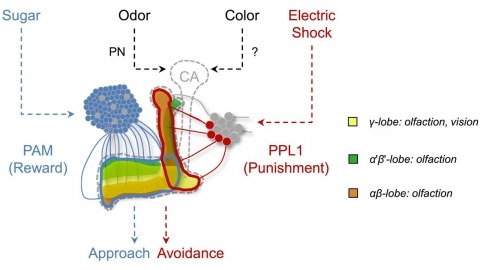
In nature, animals form memories associating reward or punishment with stimuli from different sensory modalities, such as smells and colors. It is unclear, however, how distinct sensory memories are processed in the brain. We established appetitive and aversive visual learning assays for Drosophila that are comparable to the widely used olfactory learning assays. These assays share critical features, such as reinforcing stimuli (sugar reward and electric shock punishment), and allow direct comparison of the cellular requirements for visual and olfactory memories. We found that the same subsets of dopamine neurons drive formation of both sensory memories. Furthermore, distinct yet partially overlapping subsets of mushroom body intrinsic neurons are required for visual and olfactory memories. Thus, our results suggest that distinct sensory memories are processed in a common brain center. Such centralization of related brain functions is an economical design that avoids the repetition of similar circuit motifs.
An important strategy for efficient neural coding is to match the range of cellular responses to the distribution of relevant input signals. However, the structure and relevance of sensory signals depend on behavioral state. Here, we show that behavior modifies neural activity at the earliest stages of fly vision. We describe a class of wide-field neurons that provide feedback to the most peripheral layer of the Drosophila visual system, the lamina. Using in vivo patch-clamp electrophysiology, we found that lamina wide-field neurons respond to low-frequency luminance fluctuations. Recordings in flying flies revealed that the gain and frequency tuning of wide-field neurons change during flight, and that these effects are mimicked by the neuromodulator octopamine. Genetically silencing wide-field neurons increased behavioral responses to slow-motion stimuli. Together, these findings identify a cell type that is gated by behavior to enhance neural coding by subtracting low-frequency signals from the inputs to motion detection circuits.
Visual motion perception is critical to many animal behaviors, and flies have emerged as a powerful model system for exploring this fundamental neural computation. Although numerous studies have suggested that fly motion vision is governed by a simple neural circuit [1-3], the implementation of this circuit has remained mysterious for decades. Connectomics and neurogenetics have produced a surge in recent progress, and several studies have shown selectivity for light increments (ON) or decrements (OFF) in key elements associated with this circuit [4-7]. However, related studies have reached disparate conclusions about where this selectivity emerges and whether it plays a major role in motion vision [8-13]. To address these questions, we examined activity in the neuropil thought to be responsible for visual motion detection, the medulla, of Drosophila melanogaster in response to a range of visual stimuli using two-photon calcium imaging. We confirmed that the input neurons of the medulla, the LMCs, are not responsible for light-on and light-off selectivity. We then examined the pan-neural response of medulla neurons and found prominent selectivity for light-on and light-off in layers of the medulla associated with two anatomically derived pathways (L1/L2 associated) [14, 15]. We next examined the activity of prominent interneurons within each pathway (Mi1 and Tm1) and found that these neurons have corresponding selectivity for light-on or light-off. These results provide direct evidence that motion is computed in parallel light-on and light-off pathways, demonstrate that this selectivity emerges in neurons immediately downstream of the LMCs, and specify where crucial elements of motion computation occur.
Cellular ultrastructures for signal integration are unknown in any nervous system. The ellipsoid body (EB) of the Drosophila brain is thought to control locomotion upon integration of various modalities of sensory signals with the animal internal status. However, the expected excitatory and inhibitory input convergence that virtually all brain centres exhibit is not yet described in the EB. Based on the EB expression domains of genetic constructs from the choline acetyl transferase (Cha), glutamic acid decarboxylase (GAD) and tyrosine hydroxylase (TH) genes, we identified a new set of neurons with the characteristic ring-shaped morphology (R neurons) which are presumably cholinergic, in addition to the existing GABA-expressing neurons. The R1 morphological subtype is represented in the Cha- and TH-expressing classes. In addition, using transmission electron microscopy, we identified a novel type of synapse in the EB, which exhibits the precise array of two independent active zones over the same postsynaptic dendritic domain, that we named 'agora'. This array is compatible with a coincidence detector role, and represents ~8% of all EB synapses in Drosophila. Presumably excitatory R neurons contribute to coincident synapses. Functional silencing of EB neurons by driving genetically tetanus toxin expression either reduces walking speed or alters movement orientation depending on the targeted R neuron subset, thus revealing functional specialisations in the EB for locomotion control.