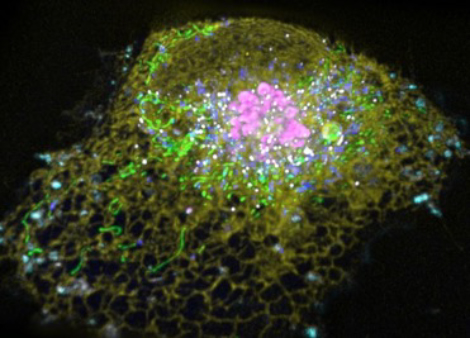
 All eukaryotic cells are comprised of similar types of subcellular organelles and cytoskeletal elements that undergo highly dynamic yet organized interactions for orchestrating complex cellular function. We are interested in understanding this active organization in the context of how it enables organ systems, comprised of diverse cell types, to carry out varied functions.
All eukaryotic cells are comprised of similar types of subcellular organelles and cytoskeletal elements that undergo highly dynamic yet organized interactions for orchestrating complex cellular function. We are interested in understanding this active organization in the context of how it enables organ systems, comprised of diverse cell types, to carry out varied functions.
To clarify the cell’s subcellular organization/dynamics and understand its significance for proper cell function in different organ cell types, we are developing and applying new multimodal imaging and analysis modalities. These include multispectral lattice light sheet imaging, grazing incidence structural illumination microscopy, single particle tracking analysis, and cryo-correlative, focused ion beam scanning electron microscopy (FIB-SEM) through entire cells. This is allowing us to better understand organelle architecture, dynamics and contact sites in healthy and diseased cell states. Our recent investigations using these and other approaches have helped define the organelle interactome (Valm et al., 2017), identified new properties of the endoplasmic reticulum (ER) and its constituents (Nixon-Abel et al., 2016), and revealed novel roles of organelle-organelle contacts (Guo et al., 2018; King et al., 2019; Chang et al., 2019), including that membrane-less RNA granules use an unusual tether, ANXA11, to couple themselves to lysosomes for long distance transport in neurons (Liao et al., 2019).
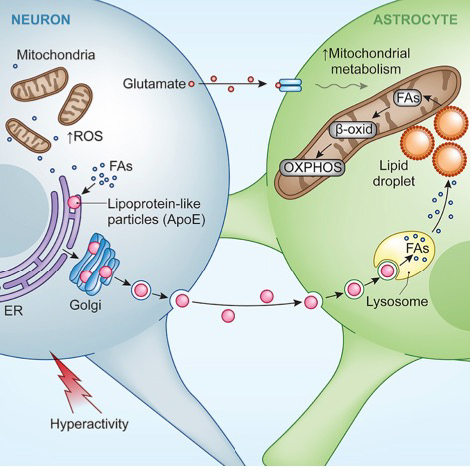
As subcellular organelles work cooperatively to drive many metabolic processes within cells, we are also examining how such cooperation occurs and how it varies among different cell types and cell states. To address this challenge, we have been combining imaging with several other techniques to explore how intracellular organelles coordinate metabolic activities to optimize function and promote survival in diverse cell types. Four cellular contexts have recently been examined: 1) glucose starvation, which we found remodels lipid trafficking pathways (Seo et al., 2021; Seo et al., 2017); 2) neuronal activity, which we discovered results in astrocytes internalizing and detoxifying damaging peroxidated lipids released as lipoprotein particles by overexcited neurons (Ioannou et al., 2019); 3) cell-cell fusion, which we found results in metabolic reprogramming towards a differentiated state (Feliciano et al., 2021), and 4) macromolecular crowding induced by osmotic change, which we discovered gives rise to nuclear liquid condensates enriched in YAP and accessible chromatin domains and leads to cell proliferation (Cai et al., 2019; 2021).
Our lab has a long-standing interest in membrane trafficking between organelles. Recently, we used cryo-correlative FIB-SEM reconstruction to investigate the first membrane trafficking event in the secretory pathway: departure from ER at ER exit sites (ERESs), demonstrating it consists of an intricate, tangled network of tubules connected to the ER by a narrow neck (Weigel et al., 2021). We are also characterizing the process of protein and lipid sorting into HIV viral particles. Here, we demonstrated using high speed TIRF analysis that such sorting relies on a lipid-based partitioning mechanism for selective protein sorting (Sengupta et al., 2019; 2020).
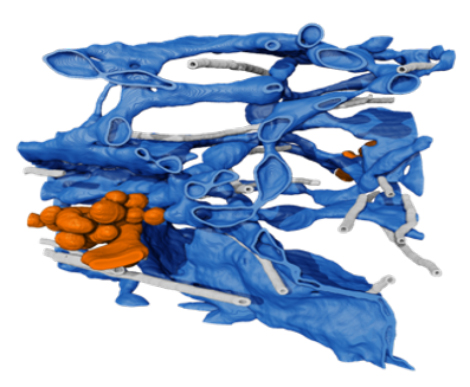
Our future investigations of organelle dynamics and interactions will increasingly be focused on cells in the context of tissues/organs as a part of Janelia’s 4D Cellular Physiology (4DCP) research direction. In this way, our group will expand its investigations of the internal states and external cues that cells negotiate from survival in a dish to their response and contribution to the function of organs and entire systems.
