We develop super-resolution methods to visualize the nano-scale organization of the 3D genome. Using lattice light-sheet microscopy and residence-time thresholding at the single-molecule level, we can systematically map stable transcription factor (TF) binding sites in single live cells (Liu et al, 2014; Xie & Dong et al, 2022). Another method, 3D ATAC-PALM, combines ATAC (Assay for Transposase-Accessible Chromatin) with lattice light-sheet PALM microscopy to selectively image nano-scale structures of the accessible genome (Xie & Peng et al, 2020). These methods, combined with chemical and genetic perturbations, allow us to investigate the molecular identities and interactions that contribute to the formation and maintenance of 3D genome organization. With our collaborators, we found that disease-inducing protein hubs promote clustering of accessible chromatin (Cai et al, 2019; Liu et al, 2023). The genome architectural protein CTCF decompacts accessible chromatin (Xie & Dong et al, 2020). In contrast, cohesin prevents spatial mixing of accessible chromatin by counterbalancing affinity interactions mediated by Brd2 (Xie & Dong et al, 2022).
Deep-tissue spatial omics imaging
By single-cell imaging and genomics, we recently discovered that 3D genome topology, mediated by cohesin, regulates gene co-expression in single cells rather than dictates population-wide gene expression levels (Dong et al, 2024).
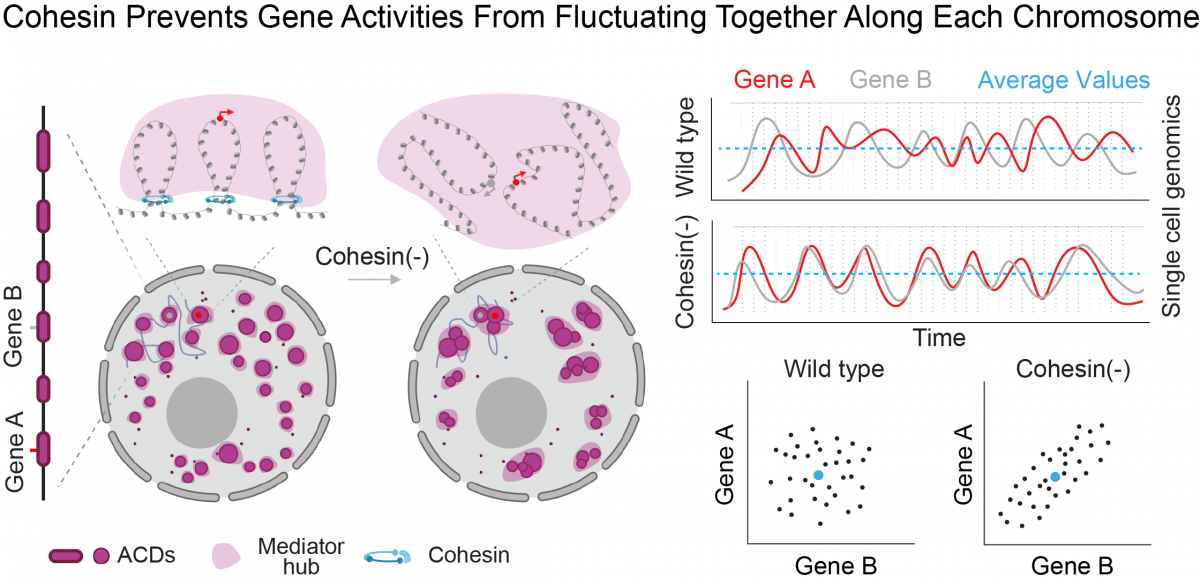
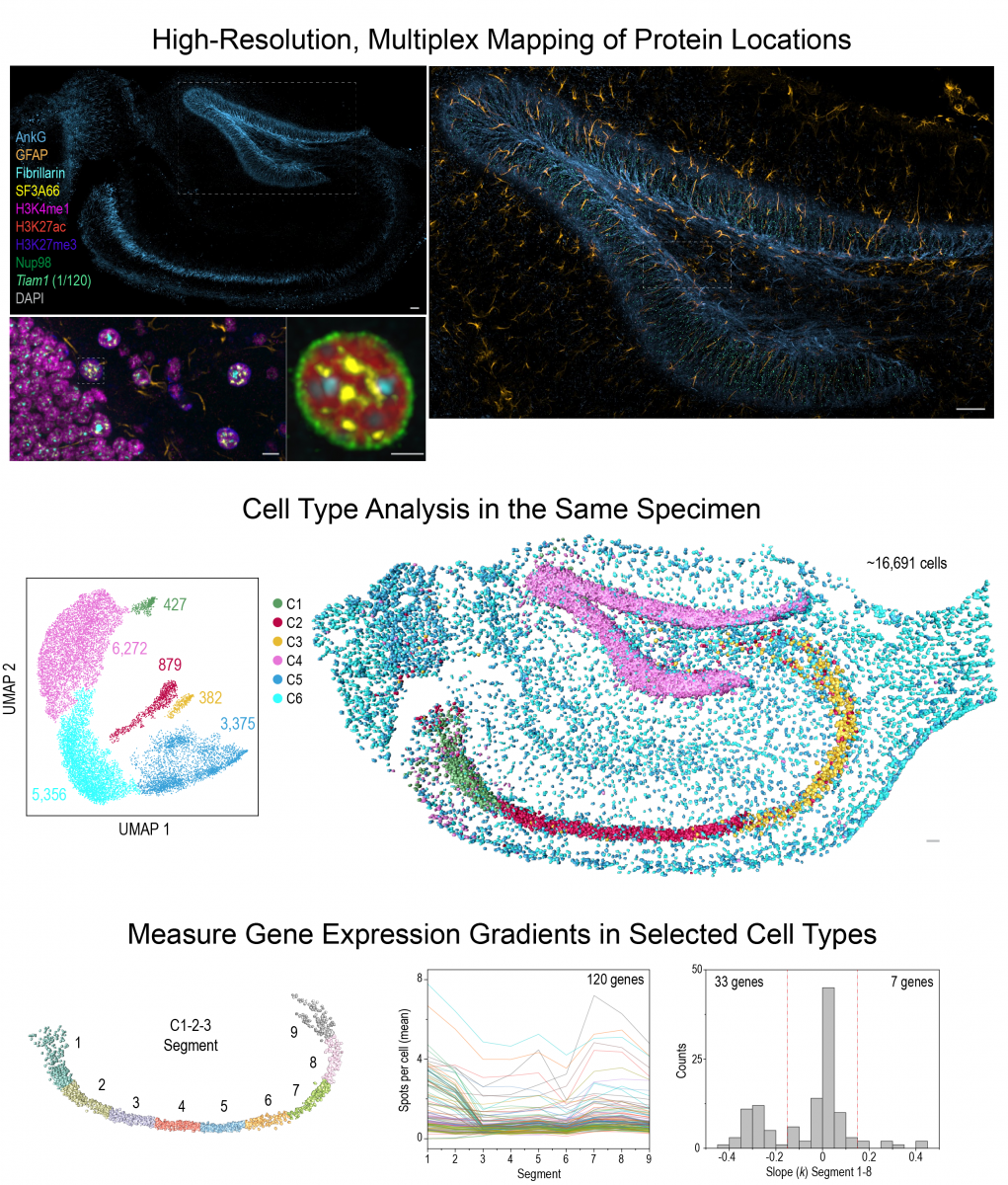
Based on this insight, we build multi-omics imaging tools (Gandin et al, 2025) to uncover the mechanisms and tissue-level functions associated with this layer of genomic regulation. Our research focuses on how high dimensional gene co-expression patterns are established during development and maintained in adult tissues. Combining multiplex RNA and protein imaging with genetic perturbations, we aim to identify molecules and networks that regulate gene co-expression and sub-cellular organization within individual cells across large tissue samples. These insights are crucial for understanding how the genome keeps cell types diverse and tissues healthy throughout an animal's life.
Imaging building blocks: molecules
We develop imaging techniques to study molecular dynamics in living cells at the single-molecule level (Chen et al, 2014; Li et al, 2016; Liu et al, 2018). We apply these techniques to examine the interplay between genome organization and transcription factor dynamics (Liu et al, 2014; Xie & Dong et al, 2022; Dong et al, 2024). We also devised new methods for molecular labeling and imaging in neurons, both in intact zebrafish and neural cultures. These approaches uncovered enhanced anterograde trafficking of synaptic vesicle precursors at the axonal initial segment (AIS) (Liu et al, 2018), lipoprotein transferring from neurons to astrocytes (Ioannou et al, 2019), and differential sorting mechanisms that target two brain enriched voltage-gated sodium channels (Nav1.2 and Nav1.6) into distinct neuronal compartments (Liu et al, 2022). In addition, we analyzed live-cell dynamics of CRSPR/Cas9 (Knight et al, 2015), transcription factors during B-cell activation (Kieffer-Kwon et al, 2017), cancer inducing TF hubs (Chong et al, 2018, Hao et al, 2024), MeCP2 and its Rett syndrome mutations (Piccolo et al, 2019), 'chemigenetic' sensor (Abdelfattah et al, 2019), ecDNAs (Hung et al, 2021; Lange et al, 2022) and CRISPR/Cas12 labeled genomic loci (Yang et al, 2024) with our collaborators.

