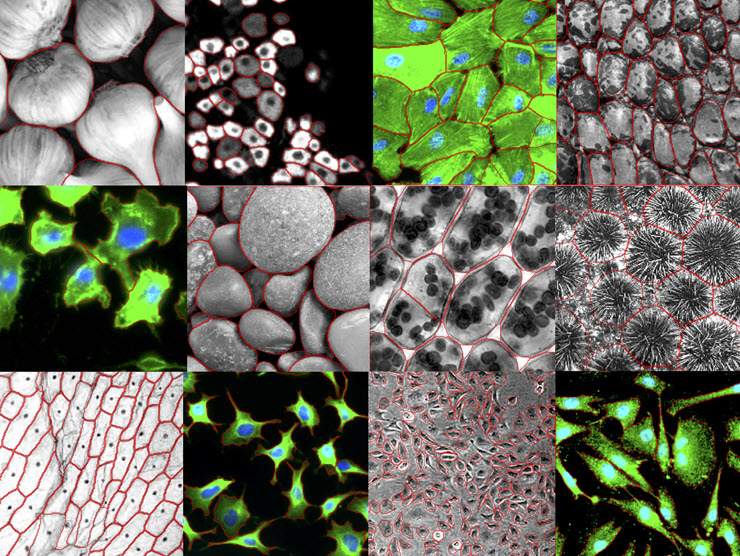
Rigid, brick-like plant cells. Elongated, tendril-bearing neurons. Round, squishy blood cells. The basic unit of life assumes many forms.
A new computer program can automatically pick out individual cells, regardless of their shape, in microscope images.
Usually, such tools are optimized for very specific data, tuned to one kind of cell with certain characteristics. But this one, called Cellpose, works far more broadly. It’s designed to segment, or map the boundaries of, many different kinds of cells. It can trace the outlines of other kinds of repeating objects, too, such as a bunch of garlic heads spilled out on a countertop. And it’s made to be user-friendly.
“You don't need to be an expert at machine learning to use it,” says Carsen Stringer, a group leader at Janelia who designed the algorithm with group leader Marius Pachitariu. Cellpose, described in a paper published in Nature Methods on on December 14, 2020, can be downloaded online for free.
Biologists who work with big datasets often rely on computers to automate some of the analysis. They might turn to algorithms that can identify individual cell boundaries in huge libraries of microscope images, for example, rather than painstakingly trace out every cell by hand. Building such a program from scratch takes time – and lots of training data to teach the computer what to look for.
“Cellpose arose out of a bit of a frustration,” Pachitariu says. His lab had sunk time into developing tools to segment particular kinds of cells. “Then other data showed up that we couldn’t run the same tool on.”
With Cellpose, Stringer and Pachitariu drew inspiration from a 2018 Data Science Bowl competition, where teams of scientists competed to create an algorithm that could identify cell nuclei in a diverse set of microscope images. But building something that worked on whole cells, rather than just their nuclei, was a bigger challenge. There’s far more variety in cell shapes, and that can be hard for a computer to contend with.
Stringer and Pachitariu first scoured the internet to collect a diverse set of microscope images. They included images filled with closely-packed cells and images with lots of blank space between cells. They included pictures of neurons, muscle cells, and plant cells. They even included images of repeating objects that weren’t cells, such as seashells.
Their colleague Michalis Michaelos, a research technician at Janelia, went through each image, tracing out the cells’ boundaries by hand. Then, Stringer and Pachitariu assigned “flow fields” to the images, turning each image into a sort of topographic map.
On a topographic map, lines denoting hills and valleys can be used to predict where water will travel through a landscape. In Cellpose, the center of each cell is treated like a valley, and the boundary of each cell is a high point. The space between cells is like a flat plateau. All of the pixels that flow into the same valley belong to the same cell. The computer uses that information to define cell boundaries in an image.
The flow-field approach allows the computer to assign pixels to specific cells even if they’re far from the cell’s center, Stringer says. Much like a stream that starts high on the slope of a mountain might drain into a river that ultimately empties into the ocean, a pixel on the tip of a dendrite might flow toward the base of the dendrite, and then in turn flow toward the center of the cell.
The researchers trained Cellpose on hundreds of flow-field representations of cell images. Then, they challenged the algorithm on new images, tracking its accuracy by measuring, pixel-by-pixel, the overlap between the hand-traced cell outlines and the algorithm’s predictions. Cellpose was able to correctly segment cells in images that didn’t resemble the training data — a task that more specialized algorithms would struggle with.
Stringer and Pachitariu have also adapted Cellpose for 3D datasets, extending the model to trace cells through multiple layers of imaging data.
Scientists can test Cellpose online before downloading the program. And they can submit their own pictures to the training dataset. As more images are incorporated, the algorithm will become more adept at handling different kinds of data, Stringer says. “We want to make this a tool that the community can keep improving.”
###
Citation
Carsen Stringer, Tim Wang, Michalis Michaelos, and Marius Pachitariu. “Cellpose: A generalist algorithm for cellular segmentation.” Nature Methods, Published online December 14, 2020. doi:10.1038/s41592-020-01018-x.
Media Contacts
Laurel Hamers
hamersl@janelia.hhmi.org




