A machine learning algorithm can automatically pick out connections between neurons in electron microscope images of the fruit fly brain.
When solving puzzles, many brains make light work. A new tool that detects connections between neurons in the fly brain has its origins in an online scientific competition.
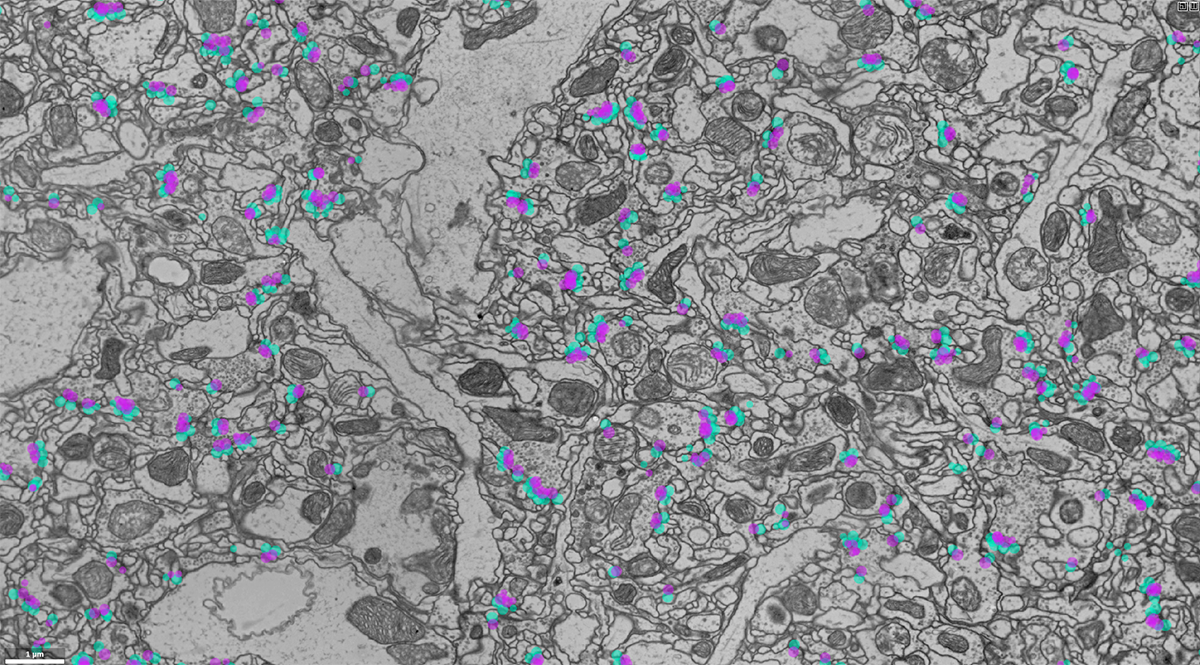
The computer program works on a dataset of 21 million electron microscope images collected at HHMI’s Janelia Research Campus—the first snapshot of the entire fruit fly brain with high enough resolution to detect individual synapses. With this tool, researchers can more efficiently use the data, mining it for precise details on fly brain circuits, the team reports June 24 in the journal Nature Methods.
The tool, which was developed by former graduate student Julia Buhmann, has detected about 250 million synaptic connections in the fruit fly brain. Finding and annotating all those synapses by hand in such a massive dataset would be nearly impossible, says group leader Jan Funke. While using automated methods to identify key features in the data can make the task manageable, developing those methods is its own challenge.
To address the gap, Funke teamed up with group leaders Stephan Saalfeld, Davi Bock, and Srini Turaga — and then turned to the broader machine learning research community.
Aided by with a team of trained annotators, Funke and his colleagues marked every single neuron and synapse in six tiny cubes pulled from different parts of the fly brain dataset. (These cubes may be small, but they’re dense enough in detail that they’d take one person a whole year to fully annotate.)
They put three of those cubes online as training data. Scientists at Janelia and other institutions competed to make the most accurate neuron tracing and synapse-detection tools, training algorithms using the hand-annotated data. The team evaluated each algorithm based on how accurately it worked on the remaining three cubes.
An algorithm developed by Buhmann rose to the top of the synapse detection leaderboard. Funke’s team then applied that algorithm to the entire dataset, creating a full map of predicted synapses in the fly brain.
Currently, the tool is more accurate in some parts of the brain than in others, Funke says, because different areas have different patterns of neural connectivity. Going forward, he hopes to improve it by training it on a more diverse dataset.
But already, the synapse-detection tool is being used by the research community. “We published our data together with a preprint describing the algorithm,” Funke says. “And because of that, several groups have already used it for their own analysis.”
###
Citation:
Julia Buhmann, Arlo Sheridan, Caroline Malin-Mayor, Philipp Schlegel, Stephan Gerhard, Tom Kazimiers, Renate Krause, Tri M. Nguyen, Larissa Heinrich, Wei-Chung Allen Lee, Rachel Wilson, Stephan Saalfeld, Gregory S. X. E. Jefferis, Davi D. Bock, Srinivas C. Turaga, Matthew Cook and Jan Funke. “Automatic detection of synaptic partners in a whole-brain Drosophila electron microscopy data set,” Nature Methods, Published online June 24, 2021. Doi:10.1038/s41592-021-01183-7




