New proteomics research is enabling scientists to decipher how neurotransmitter receptors behave and change as an organism develops. The new work could help scientists better understand the formation and function of synapses—the junctions where communication signals are passed between neurons.
In the new work, researchers in the Li Lab at Janelia and the lab of Quan Yuan at NIH characterized the proteins making up nicotinic acetylcholine receptors—the main type of neurotransmitter receptor in the fruit fly brain. These receptors, found on one side of the synapse, receive incoming signals that trigger a change in the cell’s activity.
In flies, these receptors are made up of different combinations of 10 discrete subunits, with specific configurations leading to distinct receptor functions. To understand the composition of these receptors and how they change during development, the researchers profiled the proteins in the subunits in both larva and adult flies.
They found that the fly brain utilizes different subunits as the insect develops, and that when one subunit is disrupted, another subunit compensates for the loss. The team also found that each subunit has a companion subunit, and these pairs act together: if one is disrupted, the other will also be disrupted; if one steps in to compensate, the other will also step in to compensate.
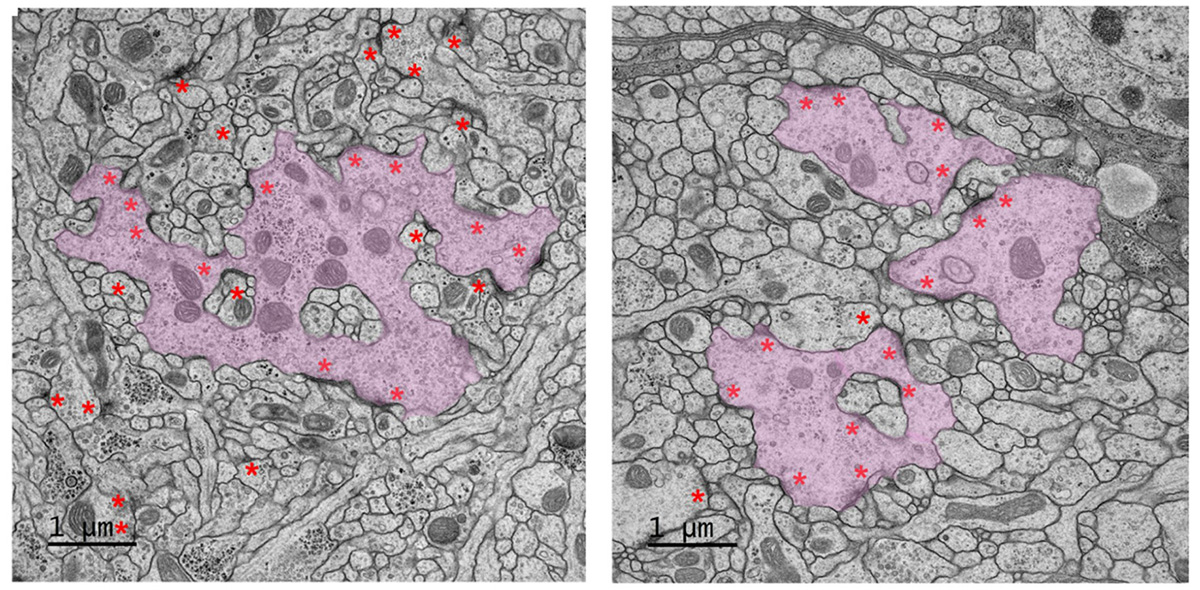
The team also identified a protein that is a key regulator of the synapse: It is present through all stages of development and, when removed, causes the entire synapse to collapse. By collaborating with the Electron Microscopy Support Team at Janelia, the researchers were able to see this collapse happening in high resolution.
Together, the new findings lay the groundwork for understanding the molecular composition, regulation, and adaptability of these synapses. Knowing how synapses form and function could give researchers new insight into the role these junctions play in aging and neurological diseases. The approach used in the new study could also be used to study different types of synapses and different organisms, helping researchers better understand the brain’s functional evolution.
###
Citation:
Justin S. Rosenthal, Dean Zhang, Jun Yin, Caixia Long, George Yang, Yan Li, Zhiyuan Lu, Wei- Ping Li, Zhiheng Yu, Jiefu Li, and Quan Yuan. “Molecular organization of central cholinergic synapses.” PMID: 40273107




