Filter
Associated Lab
- Ahrens Lab (3) Apply Ahrens Lab filter
- Aso Lab (3) Apply Aso Lab filter
- Baker Lab (5) Apply Baker Lab filter
- Betzig Lab (8) Apply Betzig Lab filter
- Branson Lab (6) Apply Branson Lab filter
- Card Lab (1) Apply Card Lab filter
- Cardona Lab (1) Apply Cardona Lab filter
- Chklovskii Lab (2) Apply Chklovskii Lab filter
- Cui Lab (2) Apply Cui Lab filter
- Dickson Lab (5) Apply Dickson Lab filter
- Druckmann Lab (1) Apply Druckmann Lab filter
- Dudman Lab (4) Apply Dudman Lab filter
- Eddy/Rivas Lab (3) Apply Eddy/Rivas Lab filter
- Egnor Lab (1) Apply Egnor Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Freeman Lab (3) Apply Freeman Lab filter
- Gonen Lab (10) Apply Gonen Lab filter
- Grigorieff Lab (3) Apply Grigorieff Lab filter
- Harris Lab (2) Apply Harris Lab filter
- Heberlein Lab (1) Apply Heberlein Lab filter
- Hess Lab (5) Apply Hess Lab filter
- Jayaraman Lab (1) Apply Jayaraman Lab filter
- Ji Lab (3) Apply Ji Lab filter
- Karpova Lab (1) Apply Karpova Lab filter
- Keller Lab (7) Apply Keller Lab filter
- Lavis Lab (7) Apply Lavis Lab filter
- Lee (Albert) Lab (4) Apply Lee (Albert) Lab filter
- Leonardo Lab (4) Apply Leonardo Lab filter
- Liu (Zhe) Lab (4) Apply Liu (Zhe) Lab filter
- Looger Lab (11) Apply Looger Lab filter
- Magee Lab (1) Apply Magee Lab filter
- Menon Lab (3) Apply Menon Lab filter
- Murphy Lab (1) Apply Murphy Lab filter
- Reiser Lab (2) Apply Reiser Lab filter
- Riddiford Lab (4) Apply Riddiford Lab filter
- Rubin Lab (8) Apply Rubin Lab filter
- Saalfeld Lab (1) Apply Saalfeld Lab filter
- Scheffer Lab (7) Apply Scheffer Lab filter
- Simpson Lab (2) Apply Simpson Lab filter
- Singer Lab (4) Apply Singer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Stern Lab (6) Apply Stern Lab filter
- Sternson Lab (5) Apply Sternson Lab filter
- Svoboda Lab (7) Apply Svoboda Lab filter
- Tervo Lab (1) Apply Tervo Lab filter
- Tjian Lab (4) Apply Tjian Lab filter
- Truman Lab (1) Apply Truman Lab filter
- Wu Lab (2) Apply Wu Lab filter
- Zlatic Lab (1) Apply Zlatic Lab filter
- Zuker Lab (1) Apply Zuker Lab filter
Associated Project Team
Associated Support Team
Publication Date
- December 2014 (31) Apply December 2014 filter
- November 2014 (7) Apply November 2014 filter
- October 2014 (14) Apply October 2014 filter
- September 2014 (14) Apply September 2014 filter
- August 2014 (11) Apply August 2014 filter
- July 2014 (23) Apply July 2014 filter
- June 2014 (13) Apply June 2014 filter
- May 2014 (10) Apply May 2014 filter
- April 2014 (18) Apply April 2014 filter
- March 2014 (13) Apply March 2014 filter
- February 2014 (11) Apply February 2014 filter
- January 2014 (25) Apply January 2014 filter
- Remove 2014 filter 2014
190 Janelia Publications
Showing 81-90 of 190 resultsThe automated tape-collecting ultramicrotome (ATUM) makes it possible to collect large numbers of ultrathin sections quickly-the equivalent of a petabyte of high resolution images each day. However, even high throughput image acquisition strategies generate images far more slowly (at present ~1 terabyte per day). We therefore developed WaferMapper, a software package that takes a multi-resolution approach to mapping and imaging select regions within a library of ultrathin sections. This automated method selects and directs imaging of corresponding regions within each section of an ultrathin section library (UTSL) that may contain many thousands of sections. Using WaferMapper, it is possible to map thousands of tissue sections at low resolution and target multiple points of interest for high resolution imaging based on anatomical landmarks. The program can also be used to expand previously imaged regions, acquire data under different imaging conditions, or re-image after additional tissue treatments.
Many biomolecules in cells can be visualized with high sensitivity and specificity by fluorescence microscopy. However, the resolution of conventional light microscopy is limited by diffraction to ~200-250nm laterally and >500nm axially. Here, we describe superresolution methods based on single-molecule localization analysis of photoswitchable fluorophores (PALM: photoactivated localization microscopy) as well as our recent three-dimensional (3D) method (iPALM: interferometric PALM) that allows imaging with a resolution better than 20nm in all three dimensions. Considerations for their implementations, applications to multicolor imaging, and a recent development that extend the imaging depth of iPALM to ~750nm are discussed. As the spatial resolution of superresolution fluorescence microscopy converges with that of electron microscopy (EM), direct imaging of the same specimen using both approaches becomes feasible. This could be particularly useful for cross validation of experiments, and thus, we also describe recent methods that were developed for correlative superresolution fluorescence and EM.
The budding yeast centromere contains Cse4, a specialized histone H3 variant. Fluorescence pulse-chase analysis of an internally tagged Cse4 reveals that it is replaced with newly synthesized molecules in S phase, remaining stably associated with centromeres thereafter. In contrast, C-terminally-tagged Cse4 is functionally impaired, showing slow cell growth, cell lethality at elevated temperatures, and extra-centromeric nuclear accumulation. Recent studies using such strains gave conflicting findings regarding the centromeric abundance and cell cycle dynamics of Cse4. Our findings indicate that internally tagged Cse4 is a better reporter of the biology of this histone variant. Furthermore, the size of centromeric Cse4 clusters was precisely mapped with a new 3D-PALM method, revealing substantial compaction during anaphase. Cse4-specific chaperone Scm3 displays steady-state, stoichiometric co-localization with Cse4 at centromeres throughout the cell cycle, while undergoing exchange with a nuclear pool. These findings suggest that a stable Cse4 nucleosome is maintained by dynamic chaperone-in-residence Scm3.DOI: http://dx.doi.org/10.7554/eLife.02203.001.
BACKGROUND: High-throughput sequencing is gradually replacing microarrays as the preferred method for studying mRNA expression levels, providing nucleotide resolution and accurately measuring absolute expression levels of almost any transcript, known or novel. However, existing microarray data from clinical, pharmaceutical, and academic settings represent valuable and often underappreciated resources, and methods for assessing and improving the quality of these data are lacking. RESULTS: To quantitatively assess the quality of microarray probes, we directly compare RNA-Seq to Agilent microarrays by processing 231 unique samples from the Allen Human Brain Atlas using RNA-Seq. Both techniques provide highly consistent, highly reproducible gene expression measurements in adult human brain, with RNA-Seq slightly outperforming microarray results overall. We show that RNA-Seq can be used as ground truth to assess the reliability of most microarray probes, remove probes with off-target effects, and scale probe intensities to match the expression levels identified by RNA-Seq. These sequencing scaled microarray intensities (SSMIs) provide more reliable, quantitative estimates of absolute expression levels for many genes when compared with unscaled intensities. Finally, we validate this result in two human cell lines, showing that linear scaling factors can be applied across experiments using the same microarray platform. CONCLUSIONS: Microarrays provide consistent, reproducible gene expression measurements, which are improved using RNA-Seq as ground truth. We expect that our strategy could be used to improve probe quality for many data sets from major existing repositories.
Iterative multi-photon adaptive compensation technique (IMPACT) has been developed for wavefront measurement and compensation in highly scattering tissues. Our previous report was largely based on the measurements of fixed tissue. Here we demonstrate the advantages of IMPACT for in vivo imaging and report the latest results. In particular, we show that IMPACT can be used for functional imaging of awake mice, and greatly improve the in vivo neuron imaging in mouse cortex at large depth (~660 microns). Moreover, IMPACT enables neuron imaging through the intact skull of adult mice, which promises noninvasive optical measurements in mouse brain.
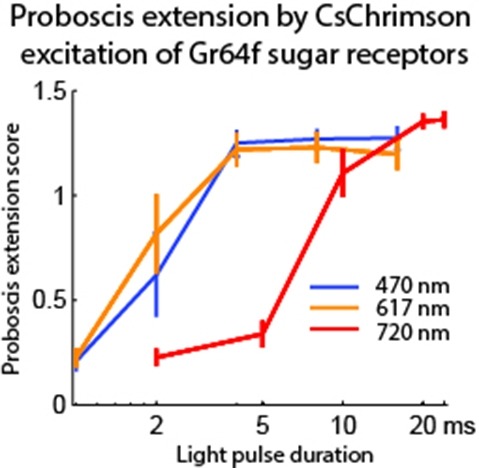
Optogenetic tools enable examination of how specific cell types contribute to brain circuit functions. A long-standing question is whether it is possible to independently activate two distinct neural populations in mammalian brain tissue. Such a capability would enable the study of how different synapses or pathways interact to encode information in the brain. Here we describe two channelrhodopsins, Chronos and Chrimson, discovered through sequencing and physiological characterization of opsins from over 100 species of alga. Chrimson’s excitation spectrum is red shifted by 45 nm relative to previous channelrhodopsins and can enable experiments in which red light is preferred. We show minimal visual system-mediated behavioral interference when using Chrimson in neurobehavioral studies in Drosophila melanogaster. Chronos has faster kinetics than previous channelrhodopsins yet is effectively more light sensitive. Together these two reagents enable two-color activation of neural spiking and downstream synaptic transmission in independent neural populations without detectable cross-talk in mouse brain slice.
The mammalian vomeronasal organ encodes pheromone information about gender, reproductive status, genetic background and individual differences. It remains unknown how pheromone information interacts to trigger innate behaviors. In this study, we identify vomeronasal receptors responsible for detecting female pheromones. A sub-group of V1re clade members recognizes gender-identifying cues in female urine. Multiple members of the V1rj clade are cognate receptors for urinary estrus signals, as well as for sulfated estrogen (SE) compounds. In both cases, the same cue activates multiple homologous receptors, suggesting redundancy in encoding female pheromone cues. Neither gender-specific cues nor SEs alone are sufficient to promote courtship behavior in male mice, whereas robust courtship behavior can be induced when the two cues are applied together. Thus, integrated action of different female cues is required in pheromone-triggered mating behavior. These results suggest a gating mechanism in the vomeronasal circuit in promoting specific innate behavior.DOI: http://dx.doi.org/10.7554/eLife.03025.001.
Sensorimotor control in vertebrates relies on internal models. When extending an arm to reach for an object, the brain uses predictive models of both limb dynamics and target properties. Whether invertebrates use such models remains unclear. Here we examine to what extent prey interception by dragonflies (Plathemis lydia), a behaviour analogous to targeted reaching, requires internal models. By simultaneously tracking the position and orientation of a dragonfly's head and body during flight, we provide evidence that interception steering is driven by forward and inverse models of dragonfly body dynamics and by models of prey motion. Predictive rotations of the dragonfly's head continuously track the prey's angular position. The head-body angles established by prey tracking appear to guide systematic rotations of the dragonfly's body to align it with the prey's flight path. Model-driven control thus underlies the bulk of interception steering manoeuvres, while vision is used for reactions to unexpected prey movements. These findings illuminate the computational sophistication with which insects construct behaviour.
In many forms of retinal degeneration, photoreceptors die but inner retinal circuits remain intact. In the rd1 mouse, an established model for blinding retinal diseases, spontaneous activity in the coupled network of AII amacrine and ON cone bipolar cells leads to rhythmic bursting of ganglion cells. Since such activity could impair retinal and/or cortical responses to restored photoreceptor function, understanding its nature is important for developing treatments of retinal pathologies. Here we analyzed a compartmental model of the wild-type mouse AII amacrine cell to predict that the cell's intrinsic membrane properties, specifically, interacting fast Na and slow, M-type K conductances, would allow its membrane potential to oscillate when light-evoked excitatory synaptic inputs were withdrawn following photoreceptor degeneration. We tested and confirmed this hypothesis experimentally by recording from AIIs in a slice preparation of rd1 retina. Additionally, recordings from ganglion cells in a whole mount preparation of rd1 retina demonstrated that activity in AIIs was propagated unchanged to elicit bursts of action potentials in ganglion cells. We conclude that oscillations are not an emergent property of a degenerated retinal network. Rather, they arise largely from the intrinsic properties of a single retinal interneuron, the AII amacrine cell.
The database iPfam, available at http://ipfam.org, catalogues Pfam domain interactions based on known 3D structures that are found in the Protein Data Bank, providing interaction data at the molecular level. Previously, the iPfam domain-domain interaction data was integrated within the Pfam database and website, but it has now been migrated to a separate database. This allows for independent development, improving data access and giving clearer separation between the protein family and interactions datasets. In addition to domain-domain interactions, iPfam has been expanded to include interaction data for domain bound small molecule ligands. Functional annotations are provided from source databases, supplemented by the incorporation of Wikipedia articles where available. iPfam (version 1.0) contains >9500 domain-domain and 15 500 domain-ligand interactions. The new website provides access to this data in a variety of ways, including interactive visualizations of the interaction data.
View Publication Page