Filter
Associated Lab
- Aso Lab (30) Apply Aso Lab filter
- Betzig Lab (1) Apply Betzig Lab filter
- Bock Lab (2) Apply Bock Lab filter
- Branson Lab (8) Apply Branson Lab filter
- Card Lab (5) Apply Card Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Dickson Lab (2) Apply Dickson Lab filter
- Druckmann Lab (1) Apply Druckmann Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Funke Lab (1) Apply Funke Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (1) Apply Heberlein Lab filter
- Hermundstad Lab (2) Apply Hermundstad Lab filter
- Hess Lab (5) Apply Hess Lab filter
- Jayaraman Lab (6) Apply Jayaraman Lab filter
- Lippincott-Schwartz Lab (1) Apply Lippincott-Schwartz Lab filter
- Looger Lab (2) Apply Looger Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Reiser Lab (15) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Romani Lab (1) Apply Romani Lab filter
- Remove Rubin Lab filter Rubin Lab
- Saalfeld Lab (4) Apply Saalfeld Lab filter
- Scheffer Lab (7) Apply Scheffer Lab filter
- Schreiter Lab (1) Apply Schreiter Lab filter
- Simpson Lab (3) Apply Simpson Lab filter
- Singer Lab (1) Apply Singer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Svoboda Lab (3) Apply Svoboda Lab filter
- Truman Lab (4) Apply Truman Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
- Turner Lab (5) Apply Turner Lab filter
Associated Project Team
- CellMap (1) Apply CellMap filter
- Fly Functional Connectome (4) Apply Fly Functional Connectome filter
- Fly Olympiad (3) Apply Fly Olympiad filter
- FlyEM (11) Apply FlyEM filter
- FlyLight (20) Apply FlyLight filter
- GENIE (1) Apply GENIE filter
- Transcription Imaging (1) Apply Transcription Imaging filter
Associated Support Team
- Project Pipeline Support (1) Apply Project Pipeline Support filter
- Electron Microscopy (4) Apply Electron Microscopy filter
- Invertebrate Shared Resource (9) Apply Invertebrate Shared Resource filter
- Janelia Experimental Technology (1) Apply Janelia Experimental Technology filter
- Management Team (1) Apply Management Team filter
- Primary & iPS Cell Culture (1) Apply Primary & iPS Cell Culture filter
- Project Technical Resources (11) Apply Project Technical Resources filter
- Quantitative Genomics (2) Apply Quantitative Genomics filter
- Scientific Computing Software (12) Apply Scientific Computing Software filter
- Scientific Computing Systems (2) Apply Scientific Computing Systems filter
Publication Date
- 2025 (4) Apply 2025 filter
- 2024 (4) Apply 2024 filter
- 2023 (5) Apply 2023 filter
- 2022 (1) Apply 2022 filter
- 2021 (4) Apply 2021 filter
- 2020 (9) Apply 2020 filter
- 2019 (6) Apply 2019 filter
- 2018 (7) Apply 2018 filter
- 2017 (15) Apply 2017 filter
- 2016 (3) Apply 2016 filter
- 2015 (16) Apply 2015 filter
- 2014 (8) Apply 2014 filter
- 2013 (5) Apply 2013 filter
- 2012 (7) Apply 2012 filter
- 2011 (3) Apply 2011 filter
- 2010 (2) Apply 2010 filter
- 2009 (1) Apply 2009 filter
- 2008 (2) Apply 2008 filter
- 2007 (2) Apply 2007 filter
- 2006 (1) Apply 2006 filter
105 Janelia Publications
Showing 101-105 of 105 resultsNervous systems combine lower-level sensory signals to detect higher-order stimulus features critical to survival, such as the visual looming motion created by an imminent collision or approaching predator. Looming-sensitive neurons have been identified in diverse animal species. Different large-scale visual features such as looming often share local cues, which means loom-detecting neurons face the challenge of rejecting confounding stimuli. Here we report the discovery of an ultra-selective looming detecting neuron, lobula plate/lobula columnar, type II (LPLC2) in Drosophila, and show how its selectivity is established by radial motion opponency. In the fly visual system, directionally selective small-field neurons called T4 and T5 form a spatial map in the lobula plate, where they each terminate in one of four retinotopic layers, such that each layer responds to motion in a different cardinal direction. Single-cell anatomical analysis reveals that each arm of the LPLC2 cross-shaped primary dendrites ramifies in one of these layers and extends along that layer's preferred motion direction. In vivo calcium imaging demonstrates that, as their shape predicts, individual LPLC2 neurons respond strongly to outward motion emanating from the centre of the neuron's receptive field. Each dendritic arm also receives local inhibitory inputs directionally selective for inward motion opposing the excitation. This radial motion opponency generates a balance of excitation and inhibition that makes LPLC2 non-responsive to related patterns of motion such as contraction, wide-field rotation or luminance change. As a population, LPLC2 neurons densely cover visual space and terminate onto the giant fibre descending neurons, which drive the jump muscle motor neuron to trigger an escape take off. Our findings provide a mechanistic description of the selective feature detection that flies use to discern and escape looming threats.
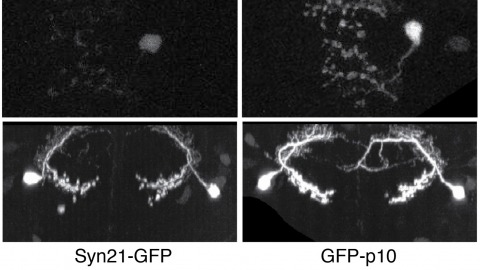
The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibire(ts1), and for the use of cytoplasmic GFP to image the fine processes of neurons.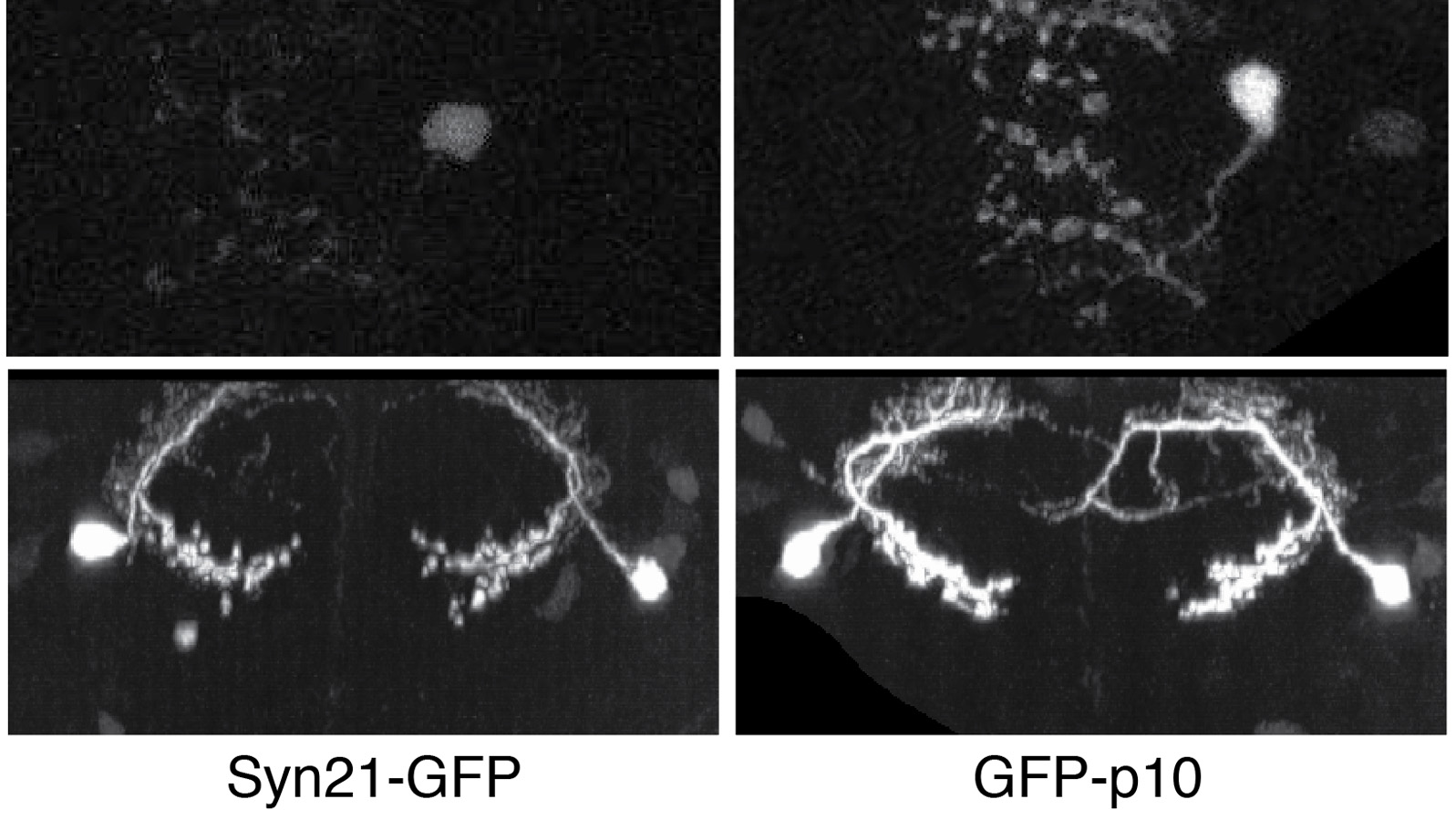
The conditional expression of hairpin constructs in Drosophila melanogaster has emerged in recent years as a method of choice in functional genomic studies. To date, upstream activating site-driven RNA interference constructs have been inserted into the genome randomly using P-element-mediated transformation, which can result in false negatives due to variable expression. To avoid this problem, we have developed a transgenic RNA interference vector based on the phiC31 site-specific integration method.
Visual projection neurons (VPNs) provide an anatomical connection between early visual processing and higher brain regions. Here we characterize lobula columnar (LC) cells, a class of Drosophila VPNs that project to distinct central brain structures called optic glomeruli. We anatomically describe 22 different LC types and show that, for several types, optogenetic activation in freely moving flies evokes specific behaviors. The activation phenotypes of two LC types closely resemble natural avoidance behaviors triggered by a visual loom. In vivo two-photon calcium imaging reveals that these LC types respond to looming stimuli, while another type does not, but instead responds to the motion of a small object. Activation of LC neurons on only one side of the brain can result in attractive or aversive turning behaviors depending on the cell type. Our results indicate that LC neurons convey information on the presence and location of visual features relevant for specific behaviors.
An important strategy for efficient neural coding is to match the range of cellular responses to the distribution of relevant input signals. However, the structure and relevance of sensory signals depend on behavioral state. Here, we show that behavior modifies neural activity at the earliest stages of fly vision. We describe a class of wide-field neurons that provide feedback to the most peripheral layer of the Drosophila visual system, the lamina. Using in vivo patch-clamp electrophysiology, we found that lamina wide-field neurons respond to low-frequency luminance fluctuations. Recordings in flying flies revealed that the gain and frequency tuning of wide-field neurons change during flight, and that these effects are mimicked by the neuromodulator octopamine. Genetically silencing wide-field neurons increased behavioral responses to slow-motion stimuli. Together, these findings identify a cell type that is gated by behavior to enhance neural coding by subtracting low-frequency signals from the inputs to motion detection circuits.