Focused ion beam scanning electron microscopy (FIB-SEM) is a technique that has been used in materials science and the semiconductor industry for multiple decades. It uses a fine atomic beam to remove a thin layer of material, then raster scans the newly exposed surface with scanning electron microscopy that uses a fine electron beam, a few nanometers in diameter, and monitors the surface by the back scattered electrons along with secondary electrons. FIB-SEM has more recently been applied in biological imaging since 2006. Biological tissues are typically stained with heavy metals, such as osmium, which binds preferentially to the cell membranes and lipids thus enhancing the electron scattering signal at such locations. After imaging, the focused Ion beam (FIB), typically comprising 30 keV gallium ions, strafes across the imaged surface and ablates a few nanometers from the top of the sample to expose a new slightly deeper surface for subsequent imaging. Cycles of etching and imaging gradually erode away the sample while enabling the collection of a stack of consecutive 2D images, usually requiring tens of seconds to a few minutes per cycle. Such continuous milling/imaging can give a stack of 3D data with fine isotropic resolution and excellent registration, also avoids many of the defects, such as tears and folds associated with cutting the thin sections required for transmission microscopy. However, the conventional FIB-SEM platform comes at the cost of slower imaging speeds and lack of long-term system stability, which in turn limit the maximum acquisition volume.
Isotropic high-resolution imaging of large volumes provides unprecedented opportunities to advance connectomics and cell biology research. With leading-edge innovations and tremendous integration effort [Xu et al. 2017; Xu et al. 2020; Xu et al. 2020], we have developed technologies to overcome the limitations of conventional systems, thereby expanding the image volume of FIB-SEM by more than four orders of magnitude from 103 µm3 to 3 x 107 µm3 while maintaining an isotropic resolution of 8 x 8 x 8 nm3 voxels. Primarily limited by time, the maximum volume can be greatly extended. These expanded volumes open a vast new regime in scientific learning, where nano-scale resolution coupled with meso and even macro scale volumes is critical. The largest connectome in the world has been generated through our enhanced FIB-SEM platform [Xu et al. 2020], where the superior z resolution empowers automated tracing of neurites and reduces the time-consuming human proofreading effort. Increased resolution further improves the interpretation of otherwise ambiguous details. Nearly all organelles can be resolved and classified with whole cell imaging at 4 nm voxel resolution. We have routinely imaged entire mammalian cells at this resolution to study the close contacts among various organelles. Furthermore, new CLEM applications [videos] enabled at the whole-cell level, can readily probe cell biology questions that are otherwise intractable.
At the forefront of volume EM imaging innovations, enhanced FIB-SEM technology pushes the envelope of image acquisition capability and system reliability, offering a novel package suited for large volume connectomics and cell biology.
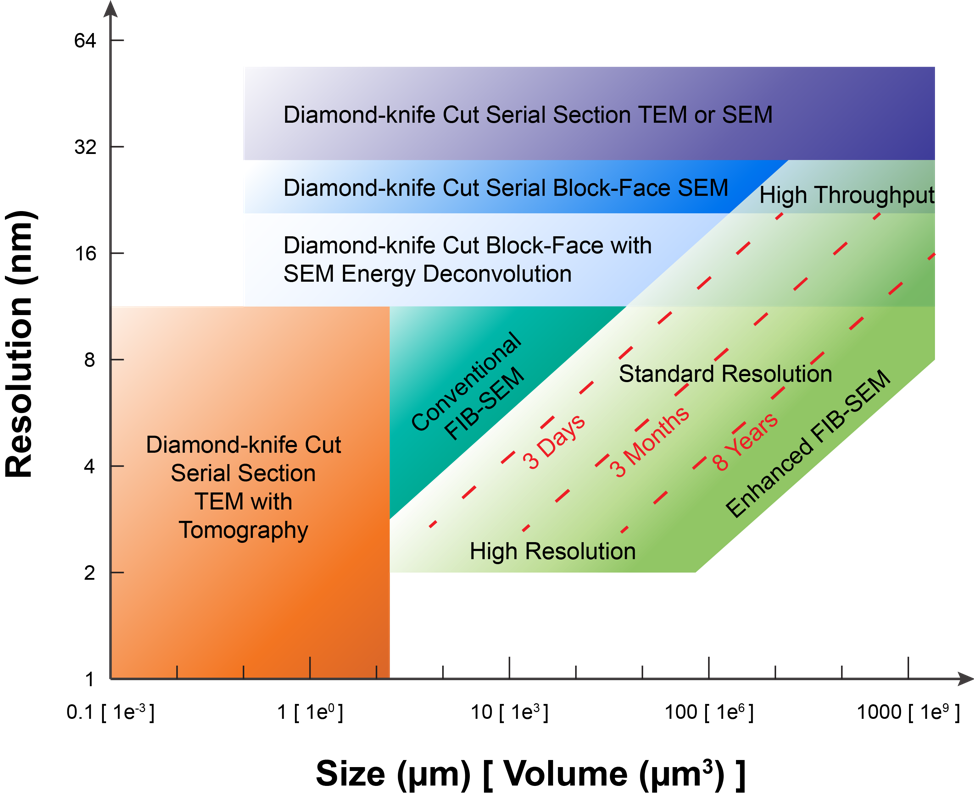
The above Figure provides an overview of the volume EM operating space. Specifically, it highlights the sample volume and minimum isotropic resolution that can be accessed only by long-term FIB-SEM imaging, and compares this to that by other volume EM imaging modalities. For example, diamond-knife cut serial section TEM with tomography, e.g. using ~500-nm-thick sections, can give better spatial resolution at the cost of imaging volume, shown at the lower left. Conversely, even larger volumes with lower spatial resolution can be collected by diamond-knife cut serial section TEM or diamond-knife cut serial block-face SEM (SBFSEM), both enclosed for reference at the top of the Figure. The space at the lower right invites a technology for connectomic studies: the fine neurites can be traced at any random orientations without degradation of resolution, while neurons expanded over long distance can be fully captured by the large volume. Our enhanced FIB-SEM system delivers a larger volume with fine spatial resolution, addressing the need for connectomic studies. To explore this FIB-SEM application space, we break the regime into three domains, labeled simply: high resolution, standard resolution, and high throughput (reduced resolution). The red diagonal dotted lines indicate contours of constant imaging time: For a given time allotted for data acquisition, one may choose either finer resolution at the expense of sample volume or vice-versa, using a single enhanced FIB-SEM system.
