Filter
Publication Date
Type of Publication
2 Publications
Showing 1-2 of 2 resultsAphid soldiers, altruistic larvae that protect the colony from predators, are an example of highly social behaviour in an insect group with a natural history different from the eusocial Hymenoptera and Isoptera. Aphids therefore allow independent tests of theory developed to explain the evolution of eusociality. Although soldiers have been discovered in five tribes from two families, the number and pattern of origins and losses of soldiers is unknown due to a lack of phylogenetic data. Here I present a mtDNA based phylogeny for the Hormaphididae, and test the hypothesis that soldiers in the tribe Cerataphidini produced during two points in the life cycle represent independent origins. The results support this hypothesis. In addition, a minimum of five evolutionary events, either four origins and one loss or five origins, are required to explain the distribution of soldiers in the family. The positions of the origins and losses are well resolved, and this phylogeny provides an historical framework for studies on the causes of soldier aphid evolution.
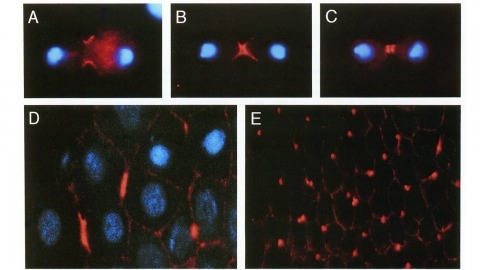
We have identified a Drosophila gene, peanut (pnut), that is related in sequence to the CDC3, CDC10, CDC11, and CDC12 genes of S. cerevisiae. These genes are required for cytokinesis, and their products are present at the bud neck during cell division. We find that pnut is also required for cytokinesis: in pnut mutants, imaginal tissues fail to proliferate and instead develop clusters of large, multinucleate cells. Pnut protein is localized to the cleavage furrow of dividing cells during cytokinesis and to the intercellular bridge connecting postmitotic daughter cells. In addition to its role in cytokinesis, pnut displays genetic interactions with seven in absentia, a gene required for neuronal fate determination in the compound eye, suggesting that pnut may have pleiotropic functions. Our results suggest that this class of proteins is involved in aspects of cytokinesis that have been conserved between flies and yeast.