Filter
Associated Lab
- Betzig Lab (4) Apply Betzig Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Gonen Lab (1) Apply Gonen Lab filter
- Harris Lab (1) Apply Harris Lab filter
- Lavis Lab (5) Apply Lavis Lab filter
- Liu (Zhe) Lab (8) Apply Liu (Zhe) Lab filter
- Rubin Lab (1) Apply Rubin Lab filter
- Singer Lab (5) Apply Singer Lab filter
- Remove Tjian Lab filter Tjian Lab
- Wu Lab (1) Apply Wu Lab filter
Associated Project Team
Publication Date
- 2016 (3) Apply 2016 filter
- 2015 (6) Apply 2015 filter
- 2014 (4) Apply 2014 filter
- 2013 (1) Apply 2013 filter
- 2012 (1) Apply 2012 filter
- 2011 (4) Apply 2011 filter
- 2010 (2) Apply 2010 filter
- 2009 (4) Apply 2009 filter
- 2008 (7) Apply 2008 filter
- 2007 (5) Apply 2007 filter
- 2006 (6) Apply 2006 filter
- 2005 (4) Apply 2005 filter
- 2004 (5) Apply 2004 filter
- 2003 (6) Apply 2003 filter
- 2002 (5) Apply 2002 filter
- 1994 (1) Apply 1994 filter
Type of Publication
64 Publications
Showing 51-60 of 64 resultsTranscriptional mechanisms that govern cellular differentiation typically include sequence-specific DNA-binding proteins and chromatin-modifying activities. These regulatory factors are assumed necessary and sufficient to drive both divergent programs of proliferation and terminal differentiation. By contrast, potential contributions of the basal transcriptional apparatus to orchestrate cell-specific gene expression have been poorly explored. In order to probe alternative mechanisms that control differentiation, we have assessed the fate of the core promoter recognition complex, TFIID, during skeletal myogenesis. Here we report that differentiation of myoblast to myotubes involves the disruption of the canonical holo-TFIID and replacement by a novel TRF3/TAF3 (TBP-related factor 3/TATA-binding protein-associated factor 3) complex. This required switching of core promoter complexes provides organisms a simple yet effective means to selectively turn on one transcriptional program while silencing many others. Although this drastic but parsimonious transcriptional switch had previously escaped our attention, it may represent a more general mechanism for regulating cell type-specific terminal differentiation.
Activator-dependent recruitment of TFIID initiates formation of the transcriptional preinitiation complex. TFIID binds core promoter DNA elements and directs the assembly of other general transcription factors, leading to binding of RNA polymerase II and activation of RNA synthesis. How TATA box-binding protein (TBP) and the TBP-associated factors (TAFs) are assembled into a functional TFIID complex with promoter recognition and coactivator activities in vivo remains unknown. Here, we use RNAi to knock down specific TFIID subunits in Drosophila tissue culture cells to determine which subunits are most critical for maintaining stability of TFIID in vivo. Contrary to expectations, we find that TAF4 rather than TBP or TAF1 plays the most critical role in maintaining stability of the complex. Our analysis also indicates that TAF5, TAF6, TAF9, and TAF12 play key roles in stability of the complex, whereas TBP, TAF1, TAF2, and TAF11 contribute very little to complex stability. Based on our results, we propose that holo-TFIID comprises a stable core subcomplex containing TAF4, TAF5, TAF6, TAF9, and TAF12 decorated with peripheral subunits TAF1, TAF2, TAF11, and TBP. Our initial functional studies indicate a specific and significant role for TAF1 and TAF4 in mediating transcription from a TATA-less, downstream core promoter element (DPE)-containing promoter, whereas a TATA-containing, DPE-less promoter was far less dependent on these subunits. In contrast to both TAF1 and TAF4, RNAi knockdown of TAF5 had little effect on transcription from either class of promoter. These studies significantly alter previous models for the assembly, structure, and function of TFIID.
With increasingly detailed images of nuclear structures revealed by advanced microscopy, a remarkably compartmentalized cell nucleus has come into focus. Although this complex nuclear organization remains largely unexplored, some progress has been made in deciphering the functional aspects of various subnuclear structures, revealing how this elaborate framework can influence gene activation. Several recent studies have helped illustrate how cells might utilize the nuclear architecture as an additional level of transcriptional control, perhaps by targeting genes and regulatory factors to specific sites within the nucleus that are designated for active RNA synthesis.
The RNA polymerase II core promoter is a structurally and functionally diverse transcriptional module. RNAi depletion and overexpression experiments revealed a genetic circuit that controls the balance of transcription from two core promoter motifs, the TATA box and the downstream core promoter element (DPE). In this circuit, TBP activates TATA-dependent transcription and represses DPE-dependent transcription, whereas Mot1 and NC2 block TBP function and thus repress TATA-dependent transcription and activate DPE-dependent transcription. This regulatory circuit is likely to be one means by which biological networks can transmit transcriptional signals, such as those from DPE-specific and TATA-specific enhancers, via distinct pathways.
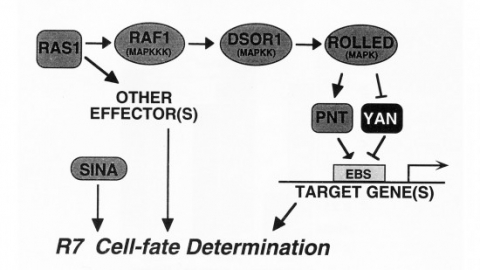
We show that the activities of two Ets-related transcription factors required for normal eye development in Drosophila, pointed and yan, are regulated by the Ras1/MAPK pathway. The pointed gene codes for two related proteins, and we show that one form is a constitutive activator of transcription, while the activity of the other form is stimulated by the Ras1/MAPK pathway. Mutation of the single consensus MAPK phosphorylation site in the second form abrogates this responsiveness. yan is a negative regulator of photoreceptor determination, and genetic data suggest that it acts as an antagonist of Ras1. We demonstrate that yan can repress transcription and that this repression activity is negatively regulated by the Ras1/MAPK signal, most likely through direct phosphorylation of yan by MAPK.
The Drosophila nucleosome remodeling factor (NURF) is an ISWI-containing chromatin remodeling complex that catalyzes ATP-dependent nucleosome sliding. By sliding nucleosomes, NURF has the ability to alter chromatin structure and regulate transcription. Previous studies have shown that mutation of Drosophila NURF induces melanotic tumors, implicating NURF in innate immune function. Here, we show that NURF mutants exhibit identical innate immune responses to gain-of-function mutants in the Drosophila JAK/STAT pathway. Using microarrays, we identify a common set of target genes that are activated in both mutants. In silico analysis of promoter sequences of these defines a consensus regulatory element comprising a STAT-binding sequence overlapped by a binding-site for the transcriptional repressor Ken. NURF interacts physically and genetically with Ken. Chromatin immunoprecipitation (ChIP) localizes NURF to Ken-binding sites in hemocytes, suggesting that Ken recruits NURF to repress STAT responders. Loss of NURF leads to precocious activation of STAT target genes.
Forty years of classical biochemical analysis have identified the molecular players involved in initiation of transcription by eukaryotic RNA polymerase II (Pol II) and largely assigned their functions. However, a dynamic picture of Pol II transcription initiation and an understanding of the mechanisms of its regulation have remained elusive due in part to inherent limitations of conventional ensemble biochemistry. Here we have begun to dissect promoter-specific transcription initiation directed by a reconstituted human Pol II system at single-molecule resolution using fluorescence video-microscopy. We detected several stochastic rounds of human Pol II transcription from individual DNA templates, observed attenuation of transcription by promoter mutations, observed enhancement of transcription by activator Sp1, and correlated the transcription signals with real-time interactions of holo-TFIID molecules at individual DNA templates. This integrated single-molecule methodology should be applicable to studying other complex biological processes.
The 100 copies of tandemly arrayed Drosophila linker (H1) and core (H2A/B and H3/H4) histone gene cluster are coordinately regulated during the cell cycle. However, the molecular mechanisms that must allow differential transcription of linker versus core histones prevalent during development remain elusive. Here, we used fluorescence imaging, biochemistry, and genetics to show that TBP (TATA-box-binding protein)-related factor 2 (TRF2) selectively regulates the TATA-less Histone H1 gene promoter, while TBP/TFIID targets core histone transcription. Importantly, TRF2-depleted polytene chromosomes display severe chromosomal structural defects. This selective usage of TRF2 and TBP provides a novel mechanism to differentially direct transcription within the histone cluster. Moreover, genome-wide chromatin immunoprecipitation (ChIP)-on-chip analyses coupled with RNA interference (RNAi)-mediated functional studies revealed that TRF2 targets several classes of TATA-less promoters of >1000 genes including those driving transcription of essential chromatin organization and protein synthesis genes. Our studies establish that TRF2 promoter recognition complexes play a significantly more central role in governing metazoan transcription than previously appreciated.
Whole-genome sequence assemblies are now available for seven different animals, including nematode worms, mice and humans. Comparative genome analyses reveal a surprising constancy in genetic content: vertebrate genomes have only about twice the number of genes that invertebrate genomes have, and the increase is primarily due to the duplication of existing genes rather than the invention of new ones. How, then, has evolutionary diversity arisen? Emerging evidence suggests that organismal complexity arises from progressively more elaborate regulation of gene expression.