Filter
Associated Lab
- Aguilera Castrejon Lab (15) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (11) Apply Ahrens Lab filter
- Baker Lab (19) Apply Baker Lab filter
- Betzig Lab (12) Apply Betzig Lab filter
- Beyene Lab (5) Apply Beyene Lab filter
- Bock Lab (3) Apply Bock Lab filter
- Branson Lab (3) Apply Branson Lab filter
- Card Lab (6) Apply Card Lab filter
- Cardona Lab (19) Apply Cardona Lab filter
- Chklovskii Lab (3) Apply Chklovskii Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Darshan Lab (4) Apply Darshan Lab filter
- Dennis Lab (1) Apply Dennis Lab filter
- Dickson Lab (14) Apply Dickson Lab filter
- Druckmann Lab (4) Apply Druckmann Lab filter
- Dudman Lab (12) Apply Dudman Lab filter
- Egnor Lab (7) Apply Egnor Lab filter
- Espinosa Medina Lab (4) Apply Espinosa Medina Lab filter
- Fetter Lab (10) Apply Fetter Lab filter
- Fitzgerald Lab (13) Apply Fitzgerald Lab filter
- Gonen Lab (32) Apply Gonen Lab filter
- Grigorieff Lab (28) Apply Grigorieff Lab filter
- Harris Lab (10) Apply Harris Lab filter
- Heberlein Lab (81) Apply Heberlein Lab filter
- Hermundstad Lab (4) Apply Hermundstad Lab filter
- Hess Lab (3) Apply Hess Lab filter
- Jayaraman Lab (4) Apply Jayaraman Lab filter
- Johnson Lab (5) Apply Johnson Lab filter
- Kainmueller Lab (19) Apply Kainmueller Lab filter
- Karpova Lab (1) Apply Karpova Lab filter
- Keleman Lab (5) Apply Keleman Lab filter
- Keller Lab (15) Apply Keller Lab filter
- Koay Lab (16) Apply Koay Lab filter
- Lavis Lab (12) Apply Lavis Lab filter
- Lee (Albert) Lab (5) Apply Lee (Albert) Lab filter
- Leonardo Lab (4) Apply Leonardo Lab filter
- Li Lab (24) Apply Li Lab filter
- Lippincott-Schwartz Lab (72) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (5) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (5) Apply Liu (Zhe) Lab filter
- Looger Lab (1) Apply Looger Lab filter
- Magee Lab (18) Apply Magee Lab filter
- Menon Lab (6) Apply Menon Lab filter
- Murphy Lab (7) Apply Murphy Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (12) Apply Otopalik Lab filter
- Pachitariu Lab (12) Apply Pachitariu Lab filter
- Pastalkova Lab (13) Apply Pastalkova Lab filter
- Pavlopoulos Lab (12) Apply Pavlopoulos Lab filter
- Pedram Lab (11) Apply Pedram Lab filter
- Reiser Lab (6) Apply Reiser Lab filter
- Riddiford Lab (24) Apply Riddiford Lab filter
- Romani Lab (12) Apply Romani Lab filter
- Rubin Lab (38) Apply Rubin Lab filter
- Saalfeld Lab (17) Apply Saalfeld Lab filter
- Satou Lab (15) Apply Satou Lab filter
- Schreiter Lab (17) Apply Schreiter Lab filter
- Sgro Lab (20) Apply Sgro Lab filter
- Simpson Lab (5) Apply Simpson Lab filter
- Singer Lab (43) Apply Singer Lab filter
- Spruston Lab (36) Apply Spruston Lab filter
- Stern Lab (83) Apply Stern Lab filter
- Sternson Lab (7) Apply Sternson Lab filter
- Stringer Lab (3) Apply Stringer Lab filter
- Svoboda Lab (4) Apply Svoboda Lab filter
- Tebo Lab (24) Apply Tebo Lab filter
- Tillberg Lab (3) Apply Tillberg Lab filter
- Tjian Lab (47) Apply Tjian Lab filter
- Truman Lab (30) Apply Truman Lab filter
- Turaga Lab (12) Apply Turaga Lab filter
- Turner Lab (11) Apply Turner Lab filter
- Wang (Shaohe) Lab (19) Apply Wang (Shaohe) Lab filter
- Wu Lab (1) Apply Wu Lab filter
- Zlatic Lab (2) Apply Zlatic Lab filter
- Zuker Lab (20) Apply Zuker Lab filter
Associated Project Team
Publication Date
- 2024 (1) Apply 2024 filter
- 2023 (1) Apply 2023 filter
- 2022 (26) Apply 2022 filter
- 2021 (19) Apply 2021 filter
- 2020 (19) Apply 2020 filter
- 2019 (25) Apply 2019 filter
- 2018 (26) Apply 2018 filter
- 2017 (31) Apply 2017 filter
- 2016 (18) Apply 2016 filter
- 2015 (57) Apply 2015 filter
- 2014 (46) Apply 2014 filter
- 2013 (58) Apply 2013 filter
- 2012 (78) Apply 2012 filter
- 2011 (92) Apply 2011 filter
- 2010 (100) Apply 2010 filter
- 2009 (102) Apply 2009 filter
- 2008 (100) Apply 2008 filter
- 2007 (85) Apply 2007 filter
- 2006 (89) Apply 2006 filter
- 2005 (67) Apply 2005 filter
- 2004 (57) Apply 2004 filter
- 2003 (58) Apply 2003 filter
- 2002 (39) Apply 2002 filter
- 2001 (28) Apply 2001 filter
- 2000 (29) Apply 2000 filter
- 1999 (14) Apply 1999 filter
- 1998 (18) Apply 1998 filter
- 1997 (16) Apply 1997 filter
- 1996 (10) Apply 1996 filter
- 1995 (18) Apply 1995 filter
- 1994 (12) Apply 1994 filter
- 1993 (10) Apply 1993 filter
- 1992 (6) Apply 1992 filter
- 1991 (11) Apply 1991 filter
- 1990 (11) Apply 1990 filter
- 1989 (6) Apply 1989 filter
- 1988 (1) Apply 1988 filter
- 1987 (7) Apply 1987 filter
- 1986 (4) Apply 1986 filter
- 1985 (5) Apply 1985 filter
- 1984 (2) Apply 1984 filter
- 1983 (2) Apply 1983 filter
- 1982 (3) Apply 1982 filter
- 1981 (3) Apply 1981 filter
- 1980 (1) Apply 1980 filter
- 1979 (1) Apply 1979 filter
- 1976 (2) Apply 1976 filter
- 1973 (1) Apply 1973 filter
- 1970 (1) Apply 1970 filter
- 1967 (1) Apply 1967 filter
Type of Publication
- Remove Non-Janelia filter Non-Janelia
1417 Publications
Showing 1371-1380 of 1417 resultsA new approach to rapid sequence comparison, basic local alignment search tool (BLAST), directly approximates alignments that optimize a measure of local similarity, the maximal segment pair (MSP) score. Recent mathematical results on the stochastic properties of MSP scores allow an analysis of the performance of this method as well as the statistical significance of alignments it generates. The basic algorithm is simple and robust; it can be implemented in a number of ways and applied in a variety of contexts including straightforward DNA and protein sequence database searches, motif searches, gene identification searches, and in the analysis of multiple regions of similarity in long DNA sequences. In addition to its flexibility and tractability to mathematical analysis, BLAST is an order of magnitude faster than existing sequence comparison tools of comparable sensitivity.
It is shown experimentally that the interaction between electrons strongly influences the chemical potential of the two-dimensional (2D) electron gas. At sufficiently low temperatures and in high magnetic fields, regions of filling factor appear where (i) the chemical potential μ diminishes with increasing carrier density, i.e., the thermodynamic density of states is negative; (ii) the derivative ∂μ/∂H (H is the magnetic field) is considerably higher than the maximum value for a noninteracting 2D electron gas. Using these results, we have estimated that the energy of the e-e interaction in Si inversion layers in a magnetic field is about 1 order of magnitude less than the classical Coulomb interaction calculated for Si metal-oxide-semiconductor field-effect transistors.
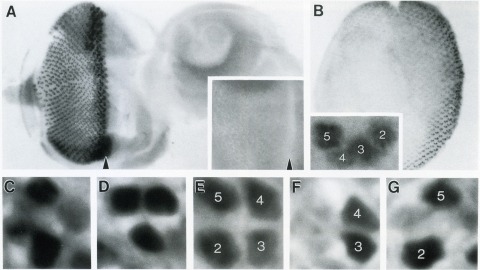
The Drosophila homeo box gene rough is required in photoreceptor cells R2 and R5 for normal eye development. We show here that rough protein expression is limited to a subset of cells in the developing retina where it is transiently expressed for 30-60 hr. The rough protein is first expressed broadly in the morphogenetic furrow but is rapidly restricted to the R2, R3, R4, and R5 precursor cells. Ubiquitous expression of rough under the control of the hsp70 promoter in third-instar larvae suppresses the initial steps of ommatidial assembly. Structures derived from other imaginal discs are not affected. Ectopic expression of rough in the R7 precursor, through the use of the sevenless promoter, causes this cell to develop into an R1-6 photoreceptor subtype; however, this cell still requires sevenless function for its neural differentiation. Taken together with previous analyses of the rough mutant phenotype, these results suggest that the normal role of rough is to establish the unique cell identity of photoreceptors R2 and R5.
The rhodopsin genes of Drosophila melanogaster are expressed in nonoverlapping subsets of photoreceptor cells within the insect visual system. Two of these genes, Rh3 and Rh4, are known to display complementary expression patterns in the UV-sensitive R7 photoreceptor cell population of the compound eye. In addition, we find that Rh3 is expressed in a small group of paired R7 and R8 photoreceptor cells at the dorsal eye margin that are apparently specialized for the detection of polarized light. In this paper we present a detailed characterization of the cis-acting requirements of both Rh3 and Rh4. Promoter deletion series demonstrate that small regulatory regions (less than 300 bp) of both R7 opsin genes contain DNA sequences sufficient to generate their respective expression patterns. Individual cis-acting elements were further identified by oligonucleotide-directed mutagenesis guided by interspecific sequence comparisons. Our results suggest that the Drosophila rhodopsin genes share a simple bipartite promoter structure, whereby the proximal region constitutes a functionally equivalent promoter "core" and the distal region determines cell-type specificity. The expression patterns of several hybrid rhodopsin promoters, in which all or part of the putative core regions have been replaced with the analogous regions of different rhodopsin promoters, provide additional evidence in support of this model.
Programmed cell death occurs in the nervous and muscular system of newly emerged adult Drosophila melanogaster. Many of the abdominal muscles that were used for eclosion and wing-spreading behavior degenerate by 12 hr after eclosion. Related neurons in the ventral ganglion also die within the first 24 hr. Ligation experiments showed that the muscle breakdown is triggered by a signal from the anterior region, presumably the head, that occurs about 1 hr before adult emergence. The timing of this signal suggests that eclosion hormone may be involved. Although muscle death is triggered prior to ecdysis, it can be delayed, at least temporarily, by forcing the emerging flies to show a prolonged ecdysis behavior. In contrast to the muscles, the death of the neurons is triggered after emergence. The signal for neuronal degeneration is closely correlated with the initiation of wing inflation behavior. Ligation and digging experiments and behavioral manipulations that either blocked or delayed wing expansion behavior had a parallel effect in suppressing or delaying neuronal death.
Transcriptional promoters of mitochondrial DNA have diverged extensively in the course of mammalian evolution. Nevertheless, the transcriptional machinery and the overall mechanisms of transcriptional control and regulation seem to be conserved. We have compared the human and murine homologs of the major DNA-binding transcriptional activator, mitochondrial transcription factor 1 (mtTF1), with unexpected results. Both proteins have similar chromatographic and transcriptional properties and are the same size. Both recognize and bind sequences between -12 and -39 within their respective homologous promoters. However, the sequences that they recognize are markedly divergent; although the base pairs they contact are situated similarly or identically with respect to the transcriptional start site, sequence identity between the two species’ contact points is less than 50%. Interestingly, the two proteins are functionally interchangeable; each can bind to the heterologous light-strand promoter and can activate transcription by the heterologous mitochondrial RNA polymerase. Thus, the RNA polymerase or some as yet undetected transcription factor, rather than mTF1, may determine the strict species specificity of mitochondrial transcription. Flexible DNA sequence recognition by mtTF1, on the other hand, may be a principal facilitating mechanism for rapid control sequence evolution.
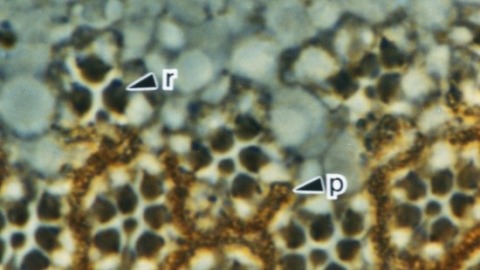
Null mutations of glass specifically remove photoreceptor cells, leaving other cell types intact. We have isolated the glass gene and have shown that its transcript encodes a putative protein of 604 amino acids with five zinc-fingers. The glass product may be a transcription factor required for the development of a single neuronal cell type.
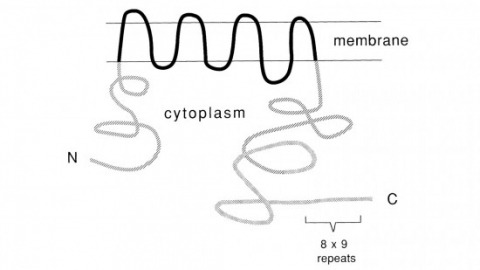
Recent studies suggest that the fly uses the inositol lipid signaling system for visual excitation and that the Drosophila transient receptor potential (trp) mutation disrupts this process subsequent to the production of IP3. In this paper, we show that trp encodes a novel 1275 amino acid protein with eight putative transmembrane segments. Immunolocalization indicates that the trp protein is expressed predominantly in the rhabdomeric membranes of the photoreceptor cells.