Filter
Associated Lab
- Aguilera Castrejon Lab (14) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (53) Apply Ahrens Lab filter
- Aso Lab (39) Apply Aso Lab filter
- Baker Lab (38) Apply Baker Lab filter
- Betzig Lab (110) Apply Betzig Lab filter
- Beyene Lab (9) Apply Beyene Lab filter
- Bock Lab (17) Apply Bock Lab filter
- Branson Lab (48) Apply Branson Lab filter
- Card Lab (38) Apply Card Lab filter
- Cardona Lab (63) Apply Cardona Lab filter
- Chklovskii Lab (13) Apply Chklovskii Lab filter
- Clapham Lab (11) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (12) Apply Darshan Lab filter
- Dennis Lab (1) Apply Dennis Lab filter
- Dickson Lab (46) Apply Dickson Lab filter
- Druckmann Lab (25) Apply Druckmann Lab filter
- Dudman Lab (46) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (11) Apply Egnor Lab filter
- Espinosa Medina Lab (16) Apply Espinosa Medina Lab filter
- Feliciano Lab (6) Apply Feliciano Lab filter
- Fetter Lab (41) Apply Fetter Lab filter
- Fitzgerald Lab (28) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (34) Apply Funke Lab filter
- Gonen Lab (91) Apply Gonen Lab filter
- Grigorieff Lab (62) Apply Grigorieff Lab filter
- Harris Lab (58) Apply Harris Lab filter
- Heberlein Lab (94) Apply Heberlein Lab filter
- Hermundstad Lab (21) Apply Hermundstad Lab filter
- Hess Lab (68) Apply Hess Lab filter
- Ilanges Lab (1) Apply Ilanges Lab filter
- Jayaraman Lab (43) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (6) Apply Johnson Lab filter
- Kainmueller Lab (19) Apply Kainmueller Lab filter
- Karpova Lab (14) Apply Karpova Lab filter
- Keleman Lab (13) Apply Keleman Lab filter
- Keller Lab (75) Apply Keller Lab filter
- Koay Lab (16) Apply Koay Lab filter
- Lavis Lab (132) Apply Lavis Lab filter
- Lee (Albert) Lab (34) Apply Lee (Albert) Lab filter
- Leonardo Lab (23) Apply Leonardo Lab filter
- Li Lab (25) Apply Li Lab filter
- Lippincott-Schwartz Lab (157) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (5) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (56) Apply Liu (Zhe) Lab filter
- Looger Lab (137) Apply Looger Lab filter
- Magee Lab (49) Apply Magee Lab filter
- Menon Lab (18) Apply Menon Lab filter
- Murphy Lab (13) Apply Murphy Lab filter
- O'Shea Lab (4) Apply O'Shea Lab filter
- Otopalik Lab (13) Apply Otopalik Lab filter
- Pachitariu Lab (40) Apply Pachitariu Lab filter
- Pastalkova Lab (18) Apply Pastalkova Lab filter
- Pavlopoulos Lab (19) Apply Pavlopoulos Lab filter
- Pedram Lab (13) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (49) Apply Reiser Lab filter
- Riddiford Lab (44) Apply Riddiford Lab filter
- Romani Lab (40) Apply Romani Lab filter
- Rubin Lab (139) Apply Rubin Lab filter
- Saalfeld Lab (58) Apply Saalfeld Lab filter
- Satou Lab (16) Apply Satou Lab filter
- Scheffer Lab (36) Apply Scheffer Lab filter
- Schreiter Lab (61) Apply Schreiter Lab filter
- Sgro Lab (20) Apply Sgro Lab filter
- Shroff Lab (21) Apply Shroff Lab filter
- Simpson Lab (23) Apply Simpson Lab filter
- Singer Lab (80) Apply Singer Lab filter
- Spruston Lab (91) Apply Spruston Lab filter
- Stern Lab (151) Apply Stern Lab filter
- Sternson Lab (54) Apply Sternson Lab filter
- Stringer Lab (26) Apply Stringer Lab filter
- Svoboda Lab (135) Apply Svoboda Lab filter
- Tebo Lab (31) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (16) Apply Tillberg Lab filter
- Tjian Lab (64) Apply Tjian Lab filter
- Truman Lab (88) Apply Truman Lab filter
- Turaga Lab (46) Apply Turaga Lab filter
- Turner Lab (35) Apply Turner Lab filter
- Vale Lab (6) Apply Vale Lab filter
- Voigts Lab (1) Apply Voigts Lab filter
- Wang (Meng) Lab (7) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (21) Apply Wang (Shaohe) Lab filter
- Wu Lab (9) Apply Wu Lab filter
- Zlatic Lab (28) Apply Zlatic Lab filter
- Zuker Lab (25) Apply Zuker Lab filter
Associated Project Team
- CellMap (1) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- Fly Descending Interneuron (10) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (50) Apply FlyEM filter
- FlyLight (46) Apply FlyLight filter
- GENIE (40) Apply GENIE filter
- Integrative Imaging (1) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (16) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (22) Apply Tool Translation Team (T3) filter
- Transcription Imaging (49) Apply Transcription Imaging filter
Publication Date
- 2024 (102) Apply 2024 filter
- 2023 (178) Apply 2023 filter
- 2022 (192) Apply 2022 filter
- 2021 (193) Apply 2021 filter
- 2020 (197) Apply 2020 filter
- 2019 (202) Apply 2019 filter
- 2018 (232) Apply 2018 filter
- 2017 (217) Apply 2017 filter
- 2016 (209) Apply 2016 filter
- 2015 (252) Apply 2015 filter
- 2014 (236) Apply 2014 filter
- 2013 (194) Apply 2013 filter
- 2012 (190) Apply 2012 filter
- 2011 (190) Apply 2011 filter
- 2010 (161) Apply 2010 filter
- 2009 (158) Apply 2009 filter
- 2008 (140) Apply 2008 filter
- 2007 (106) Apply 2007 filter
- 2006 (92) Apply 2006 filter
- 2005 (67) Apply 2005 filter
- 2004 (57) Apply 2004 filter
- 2003 (58) Apply 2003 filter
- 2002 (39) Apply 2002 filter
- 2001 (28) Apply 2001 filter
- 2000 (29) Apply 2000 filter
- 1999 (14) Apply 1999 filter
- 1998 (18) Apply 1998 filter
- 1997 (16) Apply 1997 filter
- 1996 (10) Apply 1996 filter
- 1995 (18) Apply 1995 filter
- 1994 (12) Apply 1994 filter
- 1993 (10) Apply 1993 filter
- 1992 (6) Apply 1992 filter
- 1991 (11) Apply 1991 filter
- 1990 (11) Apply 1990 filter
- 1989 (6) Apply 1989 filter
- 1988 (1) Apply 1988 filter
- 1987 (7) Apply 1987 filter
- 1986 (4) Apply 1986 filter
- 1985 (5) Apply 1985 filter
- 1984 (2) Apply 1984 filter
- 1983 (2) Apply 1983 filter
- 1982 (3) Apply 1982 filter
- 1981 (3) Apply 1981 filter
- 1980 (1) Apply 1980 filter
- 1979 (1) Apply 1979 filter
- 1976 (2) Apply 1976 filter
- 1973 (1) Apply 1973 filter
- 1970 (1) Apply 1970 filter
- 1967 (1) Apply 1967 filter
Type of Publication
3885 Publications
Showing 3861-3870 of 3885 resultsDNA primase isolated from human mitochondria sediments in glycerol density gradients at 30S and 70S. These unusually high sedimentation coefficients are a result of association of the primase activity with RNA. Treatment of primase with nuclease not only affects its sedimentation behavior, but also inactivates the primase activity. The major RNA species that cofractionates with primase activity is shown by direct sequence analysis to be cytosolic 5.8S ribosomal RNA (rRNA). Specific degradation of endogenous 5.8S rRNA using ribonuclease H and oligonucleotides complementary to 5.8S rRNA results in reduction of primase activity. Other small RNAs may play a structural role in the formation of an active DNA primase complex.
We show that the germline specificity of P element transposition is controlled at the level of mRNA splicing and not at the level of transcription. In the major P element RNA transcript, isolated from somatic cells, the first three open reading frames are joined by the removal of two introns. Using in vitro mutagenesis and genetic analysis we demonstrate the existence of a third intron whose removal is required for transposase production. We propose that this intron is only removed in the germline and that its removal is the sole basis for the germline restriction of P element transposition.
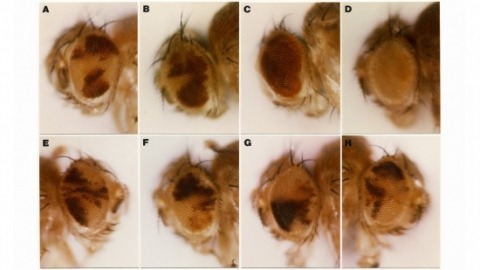
By discrete manipulation of the endocrine cues that control insect metamorphosis, it has been possible to examine the mechanisms governing the growth of neural processes during development. During the transition from larva to pupa in the hawkmoth, Manduca sexta, identified sensory neurons reorganize their central projections to evoke a new behavior–the gintrap reflex. Topical application of a juvenile hormone analog to the peripheral cell bodies of these sensory neurons during a critical period of development caused them to retain their larval commitment rather than undergo pupal development with the rest of the animal. The sensory neurons retained the larval arborization pattern within the pupal CNS and were unable to evoke the gin-trap reflex. Thus, the hormonal environment of the cell body is critical for controlling growth and synapse formation by distant axonal processes.
Synthesis of human light-strand mitochondrial DNA was accomplished in vitro using DNA primase, DNA polymerase, and other accessory proteins isolated from human mitochondria. Replication begins with the synthesis of primer RNA on a T-rich sequence in the origin stem-loop structure of the template DNA and absolutely requires ATP. A transition from RNA synthesis to DNA synthesis occurs near the base of the stem-loop structure and a potential recognition site for signaling that transition has been identified. The start sites of the in vitro products were mapped at the nucleotide level and were found to be in excellent agreement with those of in vivo nascent light-strand DNA. Isolated human mitochondrial enzymes recognize and utilize the bovine, but not the mouse, origin of light-strand replication.
Faithful transcription of human mitochondrial DNA has been reproduced in vitro, using a fraction of mitochondrial proteins capable of accurate initiation at both the heavy- and light-strand promoters. Here we report the initial dissection of this system into two nonfunctional components which, upon mixing, reconstitute promoter-specific transcriptional capacity in vitro. One of these components copurifies with the major nonspecific RNA polymerase activity, suggesting its identity. The other component lacks significant polymerase activity, but contains a protein or proteins required for accurate initiation at the two individual promoters by isolated mitochondrial RNA polymerase. This factor facilitates specific transcription, but has little or no effect on nonspecific transcription of a synthetic copolymer (poly(dA-dT)), indicating a positive role in proper promoter recognition. The transcription factor markedly stimulates light-strand transcription, but only moderately enhances transcription initiation at the heavy-strand promoter, suggesting different or additional factor requirements for heavy-strand transcription. These requirements may reflect the functional differences between heavy- and light-strand transcription in vivo and, in particular, the role of the light-strand promoter in priming of heavy-strand DNA replication.
Mammalian mitochondrial DNA maintains a novel displacement-loop region containing the major sites of transcriptional initiation and the origin of heavy strand DNA replication. Because the exact map positions of the 5’ termini of nascent mouse displacement-loop strands are known, it is possible to examine directly a potential relationship between replication priming and transcription. Analyses of in vivo nucleic acids complementary to the displacement-loop region reveal two species with identical 5’ ends at map position 16 183. One is entirely RNA and the other is RNA covalently linked to DNA. In the latter the transition from RNA to DNA is sharp, occurring near or within a series of previously identified conserved sequences 74-163 nucleotides downstream from the transcriptional initiation site. These data suggest that the initial events in replication priming and transcription are the same and that the decision to synthesize DNA or RNA is a downstream event under the control of short, conserved displacement-loop template sequences.
Using a novel method for detecting cross-homologous nucleic acid sequences we have isolated the gene coding for the major rhodopsin of Drosophila melanogaster and mapped it to chromosomal region 92B8-11. Comparison of cDNA and genomic DNA sequences indicates that the gene is divided into five exons. The amino acid sequence deduced from the nucleotide sequence is 373 residues long, and the polypeptide chain contains seven hydrophobic segments that appear to correspond to the seven transmembrane segments characteristic of other rhodopsins. Three regions of Drosophila rhodopsin are highly conserved with the corresponding domains of bovine rhodopsin, suggesting an important role for these polypeptide regions.
Many of the genes in the regulatory hierarchy controlling sex determination in Drosophila melanogaster are known. Here we examine how this regulatory hierarchy controls the expression of the structural genes encoding the female-specific yolk polypeptides. Temperature shift experiments with a temperature-sensitive allele of the sex determination regulatory gene transformer-2 (tra-2) showed that tra-2+ function is required in the adult for both the sex-specific initiation and maintenance of YP synthesis. Control of the YP genes by this regulatory hierarchy is at the level of transcription, or transcript stability. The results of temperature shift experiments with abdomens isolated from tra-2ts homozygotes support the notion that the tra-2+ function acts in a cell-autonomous manner to control YP synthesis. These results provide a paradigm for the way this regulatory hierarchy controls the terminal differentiation functions for sexually dimorphic development.
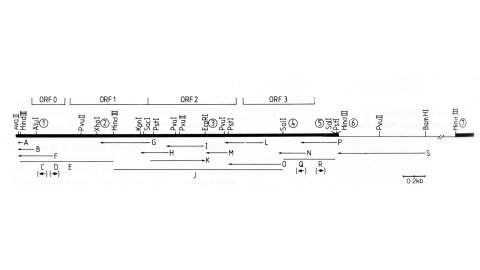
We have made a P-element derivative called Pc[ry], which carries the selectable marker gene rosy, but which acts like a nondefective, intact P element. It transposes autonomously into the germline chromosomes of an M-strain Drosophila embryo and it mobilizes in trans the defective P elements of the singed-weak allele. Frameshift mutations introduced into any of the four major open reading frames of the P sequence were each sufficient to eliminate the transposase activity, but none affected signals required in cis for transposition of the element. Complementation tests between pairs of mutant elements suggest that a single polypeptide comprises the transposase. We have examined transcripts of P elements both from natural P strains and from lines containing only nondefective Pc[ry] elements, and have identified two RNA species that appear to be specific for autonomous elements.
The major site of in vivo transcriptional initiation for both heavy and light strands of human mitochondrial DNA is the displacement-loop region. Transcripts synthesized in vitro by human mitochondrial RNA polymerase were mapped to the nucleotide level and have identical 5’ end map positions to those reported for in vivo primary transcripts. An ordered series of deletion clones, whose template sequences were truncated at either the 5’ or 3’ end, was used to identify the precise mitochondrial DNA sequence required for initiation of transcription. The data provide a definitive assignment of the promoter for heavy-strand transcription occurring within -16 to +7 of the transcriptional start site 16 nucleotides upstream of the 5’ end of the gene for tRNAPhe and of the promoter for light-strand transcription occurring within -28 to +16 of the transcriptional start site at the 5’ end of "7S RNA." Within each control sequence is a candidate promoter whose consensus sequence is 5’-CANACC(G)CC(A)AAAGAPyA-3’ and in both cases transcriptional initiation occurs within six to eight nucleotides of the 3’ end of this sequence. The transcriptional start site is an integral part of each promoter and each promoter can function in the absence of the other.