Filter
Associated Lab
- Aguilera Castrejon Lab (16) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (64) Apply Ahrens Lab filter
- Aso Lab (40) Apply Aso Lab filter
- Baker Lab (38) Apply Baker Lab filter
- Betzig Lab (112) Apply Betzig Lab filter
- Beyene Lab (13) Apply Beyene Lab filter
- Bock Lab (17) Apply Bock Lab filter
- Branson Lab (52) Apply Branson Lab filter
- Card Lab (41) Apply Card Lab filter
- Cardona Lab (63) Apply Cardona Lab filter
- Chklovskii Lab (13) Apply Chklovskii Lab filter
- Clapham Lab (14) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (12) Apply Darshan Lab filter
- Dennis Lab (1) Apply Dennis Lab filter
- Dickson Lab (46) Apply Dickson Lab filter
- Druckmann Lab (25) Apply Druckmann Lab filter
- Dudman Lab (50) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (11) Apply Egnor Lab filter
- Espinosa Medina Lab (19) Apply Espinosa Medina Lab filter
- Feliciano Lab (7) Apply Feliciano Lab filter
- Fetter Lab (41) Apply Fetter Lab filter
- Fitzgerald Lab (29) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (38) Apply Funke Lab filter
- Gonen Lab (91) Apply Gonen Lab filter
- Grigorieff Lab (62) Apply Grigorieff Lab filter
- Harris Lab (60) Apply Harris Lab filter
- Heberlein Lab (94) Apply Heberlein Lab filter
- Hermundstad Lab (26) Apply Hermundstad Lab filter
- Hess Lab (76) Apply Hess Lab filter
- Ilanges Lab (2) Apply Ilanges Lab filter
- Jayaraman Lab (46) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (6) Apply Johnson Lab filter
- Kainmueller Lab (19) Apply Kainmueller Lab filter
- Karpova Lab (14) Apply Karpova Lab filter
- Keleman Lab (13) Apply Keleman Lab filter
- Keller Lab (76) Apply Keller Lab filter
- Koay Lab (18) Apply Koay Lab filter
- Lavis Lab (148) Apply Lavis Lab filter
- Lee (Albert) Lab (34) Apply Lee (Albert) Lab filter
- Leonardo Lab (23) Apply Leonardo Lab filter
- Li Lab (28) Apply Li Lab filter
- Lippincott-Schwartz Lab (167) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (6) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (61) Apply Liu (Zhe) Lab filter
- Looger Lab (138) Apply Looger Lab filter
- Magee Lab (49) Apply Magee Lab filter
- Menon Lab (18) Apply Menon Lab filter
- Murphy Lab (13) Apply Murphy Lab filter
- O'Shea Lab (6) Apply O'Shea Lab filter
- Otopalik Lab (13) Apply Otopalik Lab filter
- Pachitariu Lab (46) Apply Pachitariu Lab filter
- Pastalkova Lab (18) Apply Pastalkova Lab filter
- Pavlopoulos Lab (19) Apply Pavlopoulos Lab filter
- Pedram Lab (15) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (51) Apply Reiser Lab filter
- Riddiford Lab (44) Apply Riddiford Lab filter
- Romani Lab (43) Apply Romani Lab filter
- Rubin Lab (143) Apply Rubin Lab filter
- Saalfeld Lab (63) Apply Saalfeld Lab filter
- Satou Lab (16) Apply Satou Lab filter
- Scheffer Lab (36) Apply Scheffer Lab filter
- Schreiter Lab (67) Apply Schreiter Lab filter
- Sgro Lab (20) Apply Sgro Lab filter
- Shroff Lab (30) Apply Shroff Lab filter
- Simpson Lab (23) Apply Simpson Lab filter
- Singer Lab (80) Apply Singer Lab filter
- Spruston Lab (93) Apply Spruston Lab filter
- Stern Lab (156) Apply Stern Lab filter
- Sternson Lab (54) Apply Sternson Lab filter
- Stringer Lab (33) Apply Stringer Lab filter
- Svoboda Lab (135) Apply Svoboda Lab filter
- Tebo Lab (33) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (21) Apply Tillberg Lab filter
- Tjian Lab (64) Apply Tjian Lab filter
- Truman Lab (88) Apply Truman Lab filter
- Turaga Lab (49) Apply Turaga Lab filter
- Turner Lab (37) Apply Turner Lab filter
- Vale Lab (7) Apply Vale Lab filter
- Voigts Lab (3) Apply Voigts Lab filter
- Wang (Meng) Lab (17) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (25) Apply Wang (Shaohe) Lab filter
- Wu Lab (9) Apply Wu Lab filter
- Zlatic Lab (28) Apply Zlatic Lab filter
- Zuker Lab (25) Apply Zuker Lab filter
Associated Project Team
- CellMap (12) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- FIB-SEM Technology (2) Apply FIB-SEM Technology filter
- Fly Descending Interneuron (10) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (53) Apply FlyEM filter
- FlyLight (49) Apply FlyLight filter
- GENIE (45) Apply GENIE filter
- Integrative Imaging (2) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (18) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (26) Apply Tool Translation Team (T3) filter
- Transcription Imaging (49) Apply Transcription Imaging filter
Publication Date
- 2025 (75) Apply 2025 filter
- 2024 (223) Apply 2024 filter
- 2023 (163) Apply 2023 filter
- 2022 (193) Apply 2022 filter
- 2021 (194) Apply 2021 filter
- 2020 (196) Apply 2020 filter
- 2019 (202) Apply 2019 filter
- 2018 (232) Apply 2018 filter
- 2017 (217) Apply 2017 filter
- 2016 (209) Apply 2016 filter
- 2015 (252) Apply 2015 filter
- 2014 (236) Apply 2014 filter
- 2013 (194) Apply 2013 filter
- 2012 (190) Apply 2012 filter
- 2011 (190) Apply 2011 filter
- 2010 (161) Apply 2010 filter
- 2009 (158) Apply 2009 filter
- 2008 (140) Apply 2008 filter
- 2007 (106) Apply 2007 filter
- 2006 (92) Apply 2006 filter
- 2005 (67) Apply 2005 filter
- 2004 (57) Apply 2004 filter
- 2003 (58) Apply 2003 filter
- 2002 (39) Apply 2002 filter
- 2001 (28) Apply 2001 filter
- 2000 (29) Apply 2000 filter
- 1999 (14) Apply 1999 filter
- 1998 (18) Apply 1998 filter
- 1997 (16) Apply 1997 filter
- 1996 (10) Apply 1996 filter
- 1995 (18) Apply 1995 filter
- 1994 (12) Apply 1994 filter
- 1993 (10) Apply 1993 filter
- 1992 (6) Apply 1992 filter
- 1991 (11) Apply 1991 filter
- 1990 (11) Apply 1990 filter
- 1989 (6) Apply 1989 filter
- 1988 (1) Apply 1988 filter
- 1987 (7) Apply 1987 filter
- 1986 (4) Apply 1986 filter
- 1985 (5) Apply 1985 filter
- 1984 (2) Apply 1984 filter
- 1983 (2) Apply 1983 filter
- 1982 (3) Apply 1982 filter
- 1981 (3) Apply 1981 filter
- 1980 (1) Apply 1980 filter
- 1979 (1) Apply 1979 filter
- 1976 (2) Apply 1976 filter
- 1973 (1) Apply 1973 filter
- 1970 (1) Apply 1970 filter
- 1967 (1) Apply 1967 filter
Type of Publication
4067 Publications
Showing 3931-3940 of 4067 resultsPhylogenetic footprinting has revealed that cis-regulatory enhancers consist of conserved DNA sequence clusters (CSCs). Currently, there is no systematic approach for enhancer discovery and analysis that takes full-advantage of the sequence information within enhancer CSCs.
Reconstructing neuron morphologies from fluorescence microscope images plays a critical role in neuroscience studies. It relies on image segmentation to produce initial masks either for further processing or final results to represent neuronal morphologies. This has been a challenging step due to the variation and complexity of noisy intensity patterns in neuron images acquired from microscopes. Whereas progresses in deep learning have brought the goal of accurate segmentation much closer to reality, creating training data for producing powerful neural networks is often laborious. To overcome the difficulty of obtaining a vast number of annotated data, we propose a novel strategy of using two-stage generative models to simulate training data with voxel-level labels. Trained upon unlabeled data by optimizing a novel objective function of preserving predefined labels, the models are able to synthesize realistic 3D images with underlying voxel labels. We showed that these synthetic images could train segmentation networks to obtain even better performance than manually labeled data. To demonstrate an immediate impact of our work, we further showed that segmentation results produced by networks trained upon synthetic data could be used to improve existing neuron reconstruction methods.
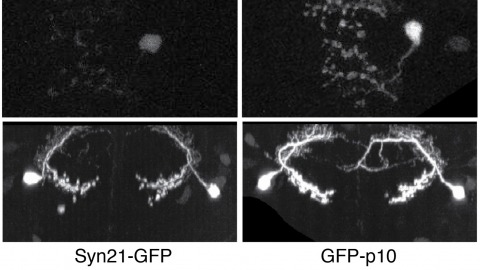
The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibire(ts1), and for the use of cytoplasmic GFP to image the fine processes of neurons.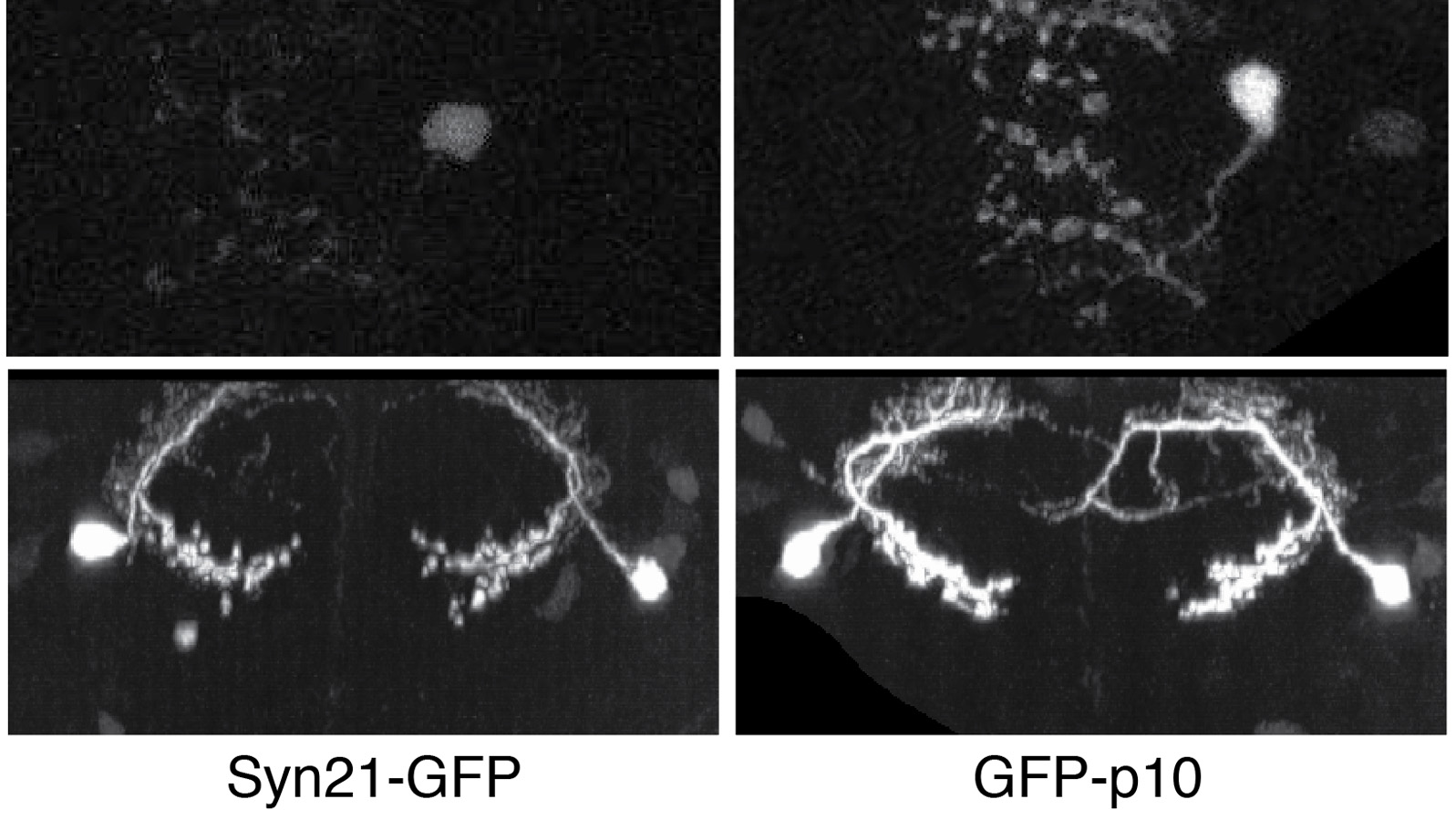
Capping Protein (CP) plays a central role in the creation of the Arp2/3-generated branched actin networks comprising lamellipodia and pseudopodia by virtue of its ability to cap the actin filament barbed end, which promotes Arp2/3-dependent filament nucleation and optimal branching. The highly conserved protein V-1/Myotrophin binds CP tightly in vitro to render it incapable of binding the barbed end. Here we addressed the physiological significance of this CP antagonist in Dictyostelium, which expresses a V-1 homolog that we show is very similar biochemically to mouse V-1. Consistent with previous studies of CP knockdown, overexpression of V-1 in Dictyostelium reduced the size of pseudopodia and the cortical content of Arp2/3 and induced the formation of filopodia. Importantly, these effects scaled positively with the degree of V-1 overexpression and were not seen with a V-1 mutant that cannot bind CP. V-1 is present in molar excess over CP, suggesting that it suppresses CP activity in the cytoplasm at steady state. Consistently, cells devoid of V-1, like cells overexpressing CP described previously, exhibited a significant decrease in cellular F-actin content. Moreover, V-1-null cells exhibited pronounced defects in macropinocytosis and chemotactic aggregation that were rescued by V-1, but not by the V-1 mutant. Together, these observations demonstrate that V-1 exerts significant influence in vivo on major actin-based processes via its ability to sequester CP. Finally, we present evidence that V-1's ability to sequester CP is regulated by phosphorylation, suggesting that cells may manipulate the level of active CP to tune their "actin phenotype."
The p2 progenitor domain in the ventral spinal cord gives rise to two interneuron subtypes: V2a and V2b. Delta-Notch-mediated cell-cell interactions between postmitotic immature neurons have been implicated in the segregation of neuron subtypes. However, lineage relationships between V2a and V2b neurons have not been reported. We address this issue using Tg[vsx1:GFP]zebrafish, a model system in which high GFP expression is initiated near the final stage of p2 progenitors. Cell fates were followed in progeny using time-lapse microscopy. Results indicate that the vast majority, if not all, of GFP-labeled p2 progenitors divide once to produce V2a/V2b neuron pairs,indicating that V2a and V2b neurons are generated by the asymmetric division of pair-producing progenitor cells. Together with evidence that Notch signaling is involved in the cell fate specification process, our results strongly suggest that Delta-Notch interactions between sister cells play a crucial role in the final outcome of these asymmetric divisions. This mechanism for determining cell fate is similar to asymmetric divisions that occur during Drosophila neurogenesis, where ganglion mother cells divide once to produce distinct neurons. However, unlike in Drosophila, the divisional axes of p2 progenitors in zebrafish were not fixed. We report that the terminal division of pair-producing progenitor cells in vertebrate neurogenesis can reproducibly produce two distinct neurons through a mechanism that may not depend on the orientation of the division axis.
The V3D system provides three-dimensional (3D) visualization of gigabyte-sized microscopy image stacks in real time on current laptops and desktops. V3D streamlines the online analysis, measurement and proofreading of complicated image patterns by combining ergonomic functions for selecting a location in an image directly in 3D space and for displaying biological measurements, such as from fluorescent probes, using the overlaid surface objects. V3D runs on all major computer platforms and can be enhanced by software plug-ins to address specific biological problems. To demonstrate this extensibility, we built a V3D-based application, V3D-Neuron, to reconstruct complex 3D neuronal structures from high-resolution brain images. V3D-Neuron can precisely digitize the morphology of a single neuron in a fruitfly brain in minutes, with about a 17-fold improvement in reliability and tenfold savings in time compared with other neuron reconstruction tools. Using V3D-Neuron, we demonstrate the feasibility of building a 3D digital atlas of neurite tracts in the fruitfly brain.
Sensory neurons innervating the lower airway provide essential feedback information that regulates respiratory physiology. These neurons synapse with second-order neurons in the central nervous system, which project directly or indirectly to the respiratory and autonomic centers. Both primary sensory neurons and second-order neurons within these circuits exhibit significant heterogeneity, and the precise roles of individual neuronal subtypes in coding the airway's internal states and modulating respiratory and autonomic outputs remain incompletely understood. In this review, we summarize recent advances in understanding the neuronal diversity along sensory circuits of the lower airway and their physiological functions. We also highlight the challenges in elucidating the roles of specific neuronal subtypes due to the extensive molecular and anatomical diversity among these neurons. Improving targeting specificity for neuronal manipulation, combined with the development of a comprehensive connectivity map, will be critical for revealing the coding and wiring logics that underlie the precise control of respiratory physiology.
Visceral sensory pathways mediate homeostatic reflexes, the dysfunction of which leads to many neurological disorders. The Bezold-Jarisch reflex (BJR), first described in 1867, is a cardioinhibitory reflex that is speculated to be mediated by vagal sensory neurons (VSNs) that also triggers syncope. However, the molecular identity, anatomical organization, physiological characteristics and behavioural influence of cardiac VSNs remain mostly unknown. Here we leveraged single-cell RNA-sequencing data and HYBRiD tissue clearing to show that VSNs that express neuropeptide Y receptor Y2 (NPY2R) predominately connect the heart ventricular wall to the area postrema. Optogenetic activation of NPY2R VSNs elicits the classic triad of BJR responses-hypotension, bradycardia and suppressed respiration-and causes an animal to faint. Photostimulation during high-resolution echocardiography and laser Doppler flowmetry with behavioural observation revealed a range of phenotypes reflected in clinical syncope, including reduced cardiac output, cerebral hypoperfusion, pupil dilation and eye-roll. Large-scale Neuropixels brain recordings and machine-learning-based modelling showed that this manipulation causes the suppression of activity across a large distributed neuronal population that is not explained by changes in spontaneous behavioural movements. Additionally, bidirectional manipulation of the periventricular zone had a push-pull effect, with inhibition leading to longer syncope periods and activation inducing arousal. Finally, ablating NPY2R VSNs specifically abolished the BJR. Combined, these results demonstrate a genetically defined cardiac reflex that recapitulates characteristics of human syncope at physiological, behavioural and neural network levels.
Volume-object annotation system (VANO) is a cross-platform image annotation system that enables one to conveniently visualize and annotate 3D volume objects including nuclei and cells. An application of VANO typically starts with an initial collection of objects produced by a segmentation computation. The objects can then be labeled, categorized, deleted, added, split, merged and redefined. VANO has been used to build high-resolution digital atlases of the nuclei of Caenorhabditis elegans at the L1 stage and the nuclei of Drosophila melanogaster’s ventral nerve cord at the late embryonic stage. AVAILABILITY: Platform independent executables of VANO, a sample dataset, and a detailed description of both its design and usage are available at research.janelia.org/peng/proj/vano. VANO is open-source for co-development.