Filter
Associated Lab
- Aso Lab (2) Apply Aso Lab filter
- Baker Lab (2) Apply Baker Lab filter
- Betzig Lab (5) Apply Betzig Lab filter
- Bock Lab (1) Apply Bock Lab filter
- Branson Lab (3) Apply Branson Lab filter
- Card Lab (1) Apply Card Lab filter
- Cardona Lab (1) Apply Cardona Lab filter
- Chklovskii Lab (1) Apply Chklovskii Lab filter
- Cui Lab (6) Apply Cui Lab filter
- Druckmann Lab (2) Apply Druckmann Lab filter
- Eddy/Rivas Lab (1) Apply Eddy/Rivas Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Gonen Lab (4) Apply Gonen Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (1) Apply Heberlein Lab filter
- Hess Lab (3) Apply Hess Lab filter
- Jayaraman Lab (2) Apply Jayaraman Lab filter
- Ji Lab (2) Apply Ji Lab filter
- Karpova Lab (1) Apply Karpova Lab filter
- Keller Lab (3) Apply Keller Lab filter
- Lavis Lab (4) Apply Lavis Lab filter
- Lee (Albert) Lab (2) Apply Lee (Albert) Lab filter
- Leonardo Lab (2) Apply Leonardo Lab filter
- Looger Lab (13) Apply Looger Lab filter
- Magee Lab (6) Apply Magee Lab filter
- Pastalkova Lab (1) Apply Pastalkova Lab filter
- Pavlopoulos Lab (1) Apply Pavlopoulos Lab filter
- Reiser Lab (1) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Rubin Lab (7) Apply Rubin Lab filter
- Saalfeld Lab (1) Apply Saalfeld Lab filter
- Scheffer Lab (3) Apply Scheffer Lab filter
- Schreiter Lab (2) Apply Schreiter Lab filter
- Simpson Lab (1) Apply Simpson Lab filter
- Spruston Lab (2) Apply Spruston Lab filter
- Sternson Lab (4) Apply Sternson Lab filter
- Svoboda Lab (9) Apply Svoboda Lab filter
- Tervo Lab (1) Apply Tervo Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Truman Lab (3) Apply Truman Lab filter
Associated Project Team
Associated Support Team
Publication Date
- December 2012 (6) Apply December 2012 filter
- November 2012 (11) Apply November 2012 filter
- October 2012 (14) Apply October 2012 filter
- September 2012 (3) Apply September 2012 filter
- August 2012 (8) Apply August 2012 filter
- July 2012 (5) Apply July 2012 filter
- June 2012 (10) Apply June 2012 filter
- May 2012 (7) Apply May 2012 filter
- April 2012 (9) Apply April 2012 filter
- March 2012 (6) Apply March 2012 filter
- February 2012 (11) Apply February 2012 filter
- January 2012 (22) Apply January 2012 filter
- Remove 2012 filter 2012
112 Janelia Publications
Showing 61-70 of 112 resultsNociception generally evokes rapid withdrawal behavior in order to protect the tissue from harmful insults. Most nociceptive neurons responding to mechanical insults display highly branched dendrites, an anatomy shared by Caenorhabditis elegans FLP and PVD neurons, which mediate harsh touch responses. Although several primary molecular nociceptive sensors have been characterized, less is known about modulation and amplification of noxious signals within nociceptor neurons. First, we analyzed the FLP/PVD network by optogenetics and studied integration of signals from these cells in downstream interneurons. Second, we investigated which genes modulate PVD function, based on prior single-neuron mRNA profiling of PVD.
Cell type-specific expression of optogenetic molecules allows temporally precise manipulation of targeted neuronal activity. Here we present a toolbox of four knock-in mouse lines engineered for strong, Cre-dependent expression of channelrhodopsins ChR2-tdTomato and ChR2-EYFP, halorhodopsin eNpHR3.0 and archaerhodopsin Arch-ER2. All four transgenes mediated Cre-dependent, robust activation or silencing of cortical pyramidal neurons in vitro and in vivo upon light stimulation, with ChR2-EYFP and Arch-ER2 demonstrating light sensitivity approaching that of in utero or virally transduced neurons. We further show specific photoactivation of parvalbumin-positive interneurons in behaving ChR2-EYFP reporter mice. The robust, consistent and inducible nature of our ChR2 mice represents a significant advance over previous lines, and the Arch-ER2 and eNpHR3.0 mice are to our knowledge the first demonstration of successful conditional transgenic optogenetic silencing. When combined with the hundreds of available Cre driver lines, this optimized toolbox of reporter mice will enable widespread investigations of neural circuit function with unprecedented reliability and accuracy.
Wide-field motion-sensitive neurons in the lobula plate (lobula plate tangential cells, LPTCs) of the fly have been studied for decades. However, it has never been conclusively shown which cells constitute their major presynaptic elements. LPTCs are supposed to be rendered directionally selective by integrating excitatory as well as inhibitory input from many local motion detectors. Based on their stratification in the different layers of the lobula plate, the columnar cells T4 and T5 are likely candidates to provide some of this input. To study their role in motion detection, we performed whole-cell recordings from LPTCs in Drosophila with T4 and T5 cells blocked using two different genetically encoded tools. In these flies, motion responses were abolished, while flicker responses largely remained. We thus demonstrate that T4 and T5 cells indeed represent those columnar cells that provide directionally selective motion information to LPTCs. Contrary to previous assumptions, flicker responses seem to be largely mediated by a third, independent pathway. This work thus represents a further step towards elucidating the complete motion detection circuitry of the fly.
A consortium of inhibitory neurons control the firing patterns of pyramidal cells, but their specific roles in the behaving animal are largely unknown. We performed simultaneous physiological recordings and optogenetic silencing of either perisomatic (parvalbumin (PV) expressing) or dendrite-targeting (somatostatin (SOM) expressing) interneurons in hippocampal area CA1 of head-fixed mice actively moving a treadmill belt rich with visual-tactile stimuli. Silencing of either PV or SOM interneurons increased the firing rates of pyramidal cells selectively in their place fields, with PV and SOM interneurons having their largest effect during the rising and decaying parts of the place field, respectively. SOM interneuron silencing powerfully increased burst firing without altering the theta phase of spikes. In contrast, PV interneuron silencing had no effect on burst firing, but instead shifted the spikes’ theta phase toward the trough of theta. These findings indicate that perisomatic and dendritic inhibition have distinct roles in controlling the rate, burst and timing of hippocampal pyramidal cells.
View Publication PageThe mechanisms linking sensation and action during learning are poorly understood. Layer 2/3 neurons in the motor cortex might participate in sensorimotor integration and learning; they receive input from sensory cortex and excite deep layer neurons, which control movement. Here we imaged activity in the same set of layer 2/3 neurons in the motor cortex over weeks, while mice learned to detect objects with their whiskers and report detection with licking. Spatially intermingled neurons represented sensory (touch) and motor behaviours (whisker movements and licking). With learning, the population-level representation of task-related licking strengthened. In trained mice, population-level representations were redundant and stable, despite dynamism of single-neuron representations. The activity of a subpopulation of neurons was consistent with touch driving licking behaviour. Our results suggest that ensembles of motor cortex neurons couple sensory input to multiple, related motor programs during learning.
Microscopic images of specific proteins in their cellular context yield important insights into biological processes and cellular architecture. The advent of superresolution optical microscopy techniques provides the possibility to augment EM with nanometer-resolution fluorescence microscopy to access the precise location of proteins in the context of cellular ultrastructure. Unfortunately, efforts to combine superresolution fluorescence and EM have been stymied by the divergent and incompatible sample preparation protocols of the two methods. Here, we describe a protocol that preserves both the delicate photoactivatable fluorescent protein labels essential for superresolution microscopy and the fine ultrastructural context of EM. This preparation enables direct 3D imaging in 500- to 750-nm sections with interferometric photoactivatable localization microscopy followed by scanning EM images generated by focused ion beam ablation. We use this process to "colorize" detailed EM images of the mitochondrion with the position of labeled proteins. The approach presented here has provided a new level of definition of the in vivo nature of organization of mitochondrial nucleoids, and we expect this straightforward method to be applicable to many other biological questions that can be answered by direct imaging.
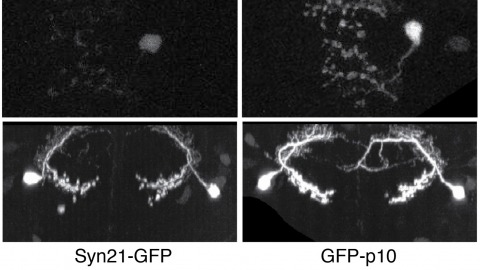
The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibire(ts1), and for the use of cytoplasmic GFP to image the fine processes of neurons.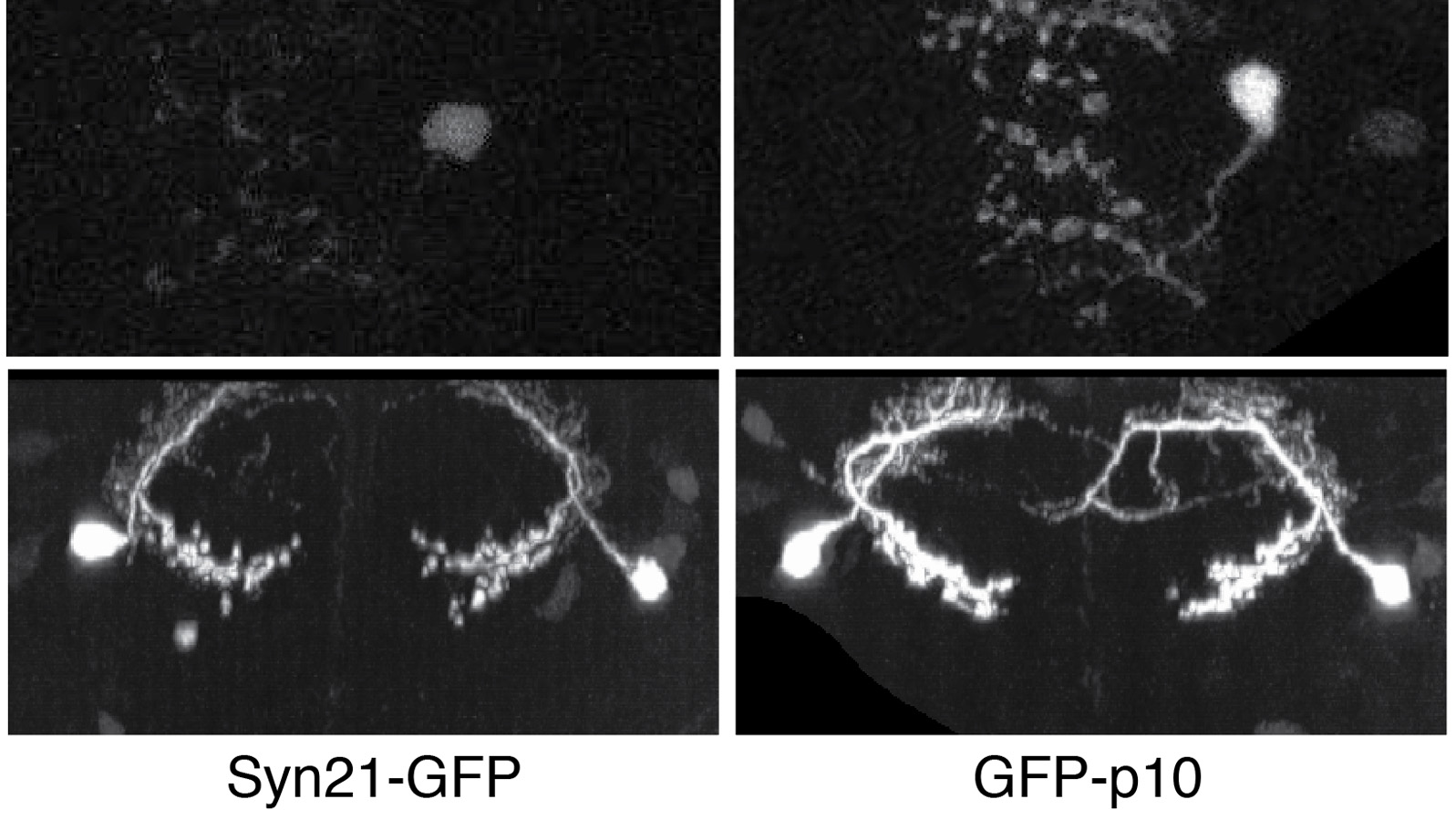
Voltage-gated ion channels are responsible for transmitting electrochemical signals in both excitable and non-excitable cells. Structural studies of voltage-gated potassium and sodium channels by X-ray crystallography have revealed atomic details on their voltage-sensor domains (VSDs) and pore domains, and were put in context of disparate mechanistic views on the voltage-driven conformational changes in these proteins. Functional investigation of voltage-gated channels in membranes, however, showcased a mechanism of lipid-dependent gating for voltage-gated channels, suggesting that the lipids play an indispensible and critical role in the proper gating of many of these channels. Structure determination of membrane-embedded voltage-gated ion channels appears to be the next frontier in fully addressing the mechanism by which the VSDs control channel opening. Currently electron crystallography is the only structural biology method in which a membrane protein of interest is crystallized within a complete lipid-bilayer mimicking the native environment of a biological membrane. At a sufficiently high resolution, an electron crystallographic structure could reveal lipids, the channel and their mutual interactions at the atomic level. Electron crystallography is therefore a promising avenue toward understanding how lipids modulate channel activation through close association with the VSDs.
Previous implementations of structured-illumination microscopy (SIM) were slow or designed for one-color excitation, sacrificing two unique and extremely beneficial aspects of light microscopy: live-cell imaging in multiple colors. This is especially unfortunate because, among the resolution-extending techniques, SIM is an attractive choice for live-cell imaging; it requires no special fluorophores or high light intensities to achieve twice diffraction-limited resolution in three dimensions. Furthermore, its wide-field nature makes it light-efficient and decouples the acquisition speed from the size of the lateral field of view, meaning that high frame rates over large volumes are possible. Here, we report a previously undescribed SIM setup that is fast enough to record 3D two-color datasets of living whole cells. Using rapidly programmable liquid crystal devices and a flexible 2D grid pattern algorithm to switch between excitation wavelengths quickly, we show volume rates as high as 4 s in one color and 8.5 s in two colors over tens of time points. To demonstrate the capabilities of our microscope, we image a variety of biological structures, including mitochondria, clathrin-coated vesicles, and the actin cytoskeleton, in either HeLa cells or cultured neurons.