Filter
Associated Lab
- Ahrens Lab (42) Apply Ahrens Lab filter
- Aso Lab (39) Apply Aso Lab filter
- Baker Lab (19) Apply Baker Lab filter
- Betzig Lab (98) Apply Betzig Lab filter
- Beyene Lab (4) Apply Beyene Lab filter
- Bock Lab (14) Apply Bock Lab filter
- Branson Lab (45) Apply Branson Lab filter
- Card Lab (32) Apply Card Lab filter
- Cardona Lab (44) Apply Cardona Lab filter
- Chklovskii Lab (10) Apply Chklovskii Lab filter
- Clapham Lab (10) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (8) Apply Darshan Lab filter
- Dickson Lab (32) Apply Dickson Lab filter
- Druckmann Lab (21) Apply Druckmann Lab filter
- Dudman Lab (34) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (4) Apply Egnor Lab filter
- Espinosa Medina Lab (12) Apply Espinosa Medina Lab filter
- Feliciano Lab (6) Apply Feliciano Lab filter
- Fetter Lab (31) Apply Fetter Lab filter
- Fitzgerald Lab (15) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (34) Apply Funke Lab filter
- Gonen Lab (59) Apply Gonen Lab filter
- Grigorieff Lab (34) Apply Grigorieff Lab filter
- Harris Lab (48) Apply Harris Lab filter
- Heberlein Lab (13) Apply Heberlein Lab filter
- Hermundstad Lab (17) Apply Hermundstad Lab filter
- Hess Lab (65) Apply Hess Lab filter
- Ilanges Lab (1) Apply Ilanges Lab filter
- Jayaraman Lab (39) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (1) Apply Johnson Lab filter
- Karpova Lab (13) Apply Karpova Lab filter
- Keleman Lab (8) Apply Keleman Lab filter
- Keller Lab (60) Apply Keller Lab filter
- Lavis Lab (120) Apply Lavis Lab filter
- Lee (Albert) Lab (29) Apply Lee (Albert) Lab filter
- Leonardo Lab (19) Apply Leonardo Lab filter
- Li Lab (1) Apply Li Lab filter
- Lippincott-Schwartz Lab (85) Apply Lippincott-Schwartz Lab filter
- Liu (Zhe) Lab (51) Apply Liu (Zhe) Lab filter
- Looger Lab (136) Apply Looger Lab filter
- Magee Lab (31) Apply Magee Lab filter
- Menon Lab (12) Apply Menon Lab filter
- Murphy Lab (6) Apply Murphy Lab filter
- O'Shea Lab (3) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Pachitariu Lab (28) Apply Pachitariu Lab filter
- Pastalkova Lab (5) Apply Pastalkova Lab filter
- Pavlopoulos Lab (7) Apply Pavlopoulos Lab filter
- Pedram Lab (2) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (43) Apply Reiser Lab filter
- Riddiford Lab (20) Apply Riddiford Lab filter
- Romani Lab (28) Apply Romani Lab filter
- Rubin Lab (101) Apply Rubin Lab filter
- Saalfeld Lab (41) Apply Saalfeld Lab filter
- Satou Lab (1) Apply Satou Lab filter
- Scheffer Lab (36) Apply Scheffer Lab filter
- Schreiter Lab (44) Apply Schreiter Lab filter
- Shroff Lab (21) Apply Shroff Lab filter
- Simpson Lab (18) Apply Simpson Lab filter
- Singer Lab (37) Apply Singer Lab filter
- Spruston Lab (55) Apply Spruston Lab filter
- Stern Lab (68) Apply Stern Lab filter
- Sternson Lab (47) Apply Sternson Lab filter
- Stringer Lab (23) Apply Stringer Lab filter
- Svoboda Lab (131) Apply Svoboda Lab filter
- Tebo Lab (7) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (13) Apply Tillberg Lab filter
- Tjian Lab (17) Apply Tjian Lab filter
- Truman Lab (58) Apply Truman Lab filter
- Turaga Lab (34) Apply Turaga Lab filter
- Turner Lab (24) Apply Turner Lab filter
- Vale Lab (6) Apply Vale Lab filter
- Voigts Lab (1) Apply Voigts Lab filter
- Wang (Meng) Lab (7) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (2) Apply Wang (Shaohe) Lab filter
- Wu Lab (8) Apply Wu Lab filter
- Zlatic Lab (26) Apply Zlatic Lab filter
- Zuker Lab (5) Apply Zuker Lab filter
Associated Project Team
- CellMap (1) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- Fly Descending Interneuron (10) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (50) Apply FlyEM filter
- FlyLight (46) Apply FlyLight filter
- GENIE (40) Apply GENIE filter
- Integrative Imaging (1) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (16) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (22) Apply Tool Translation Team (T3) filter
- Transcription Imaging (45) Apply Transcription Imaging filter
Associated Support Team
- Anatomy and Histology (18) Apply Anatomy and Histology filter
- Cryo-Electron Microscopy (33) Apply Cryo-Electron Microscopy filter
- Electron Microscopy (11) Apply Electron Microscopy filter
- Fly Facility (39) Apply Fly Facility filter
- Gene Targeting and Transgenics (10) Apply Gene Targeting and Transgenics filter
- Integrative Imaging (10) Apply Integrative Imaging filter
- Janelia Experimental Technology (35) Apply Janelia Experimental Technology filter
- Management Team (1) Apply Management Team filter
- Molecular Genomics (15) Apply Molecular Genomics filter
- Primary & iPS Cell Culture (13) Apply Primary & iPS Cell Culture filter
- Project Technical Resources (32) Apply Project Technical Resources filter
- Quantitative Genomics (18) Apply Quantitative Genomics filter
- Scientific Computing Software (56) Apply Scientific Computing Software filter
- Scientific Computing Systems (6) Apply Scientific Computing Systems filter
- Viral Tools (14) Apply Viral Tools filter
- Vivarium (6) Apply Vivarium filter
Publication Date
- 2024 (102) Apply 2024 filter
- 2023 (177) Apply 2023 filter
- 2022 (166) Apply 2022 filter
- 2021 (174) Apply 2021 filter
- 2020 (178) Apply 2020 filter
- 2019 (177) Apply 2019 filter
- 2018 (206) Apply 2018 filter
- 2017 (186) Apply 2017 filter
- 2016 (191) Apply 2016 filter
- 2015 (195) Apply 2015 filter
- 2014 (190) Apply 2014 filter
- 2013 (136) Apply 2013 filter
- 2012 (112) Apply 2012 filter
- 2011 (98) Apply 2011 filter
- 2010 (61) Apply 2010 filter
- 2009 (56) Apply 2009 filter
- 2008 (40) Apply 2008 filter
- 2007 (21) Apply 2007 filter
- 2006 (3) Apply 2006 filter
2469 Janelia Publications
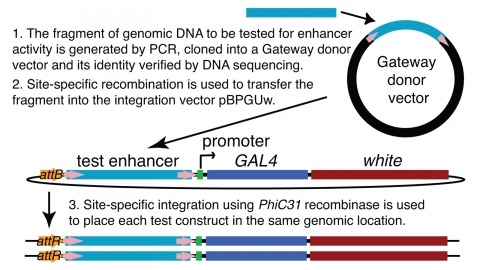
Showing 2421-2430 of 2469 resultsWe demonstrate the feasibility of generating thousands of transgenic Drosophila melanogaster lines in which the expression of an exogenous gene is reproducibly directed to distinct small subsets of cells in the adult brain. We expect the expression patterns produced by the collection of 5,000 lines that we are currently generating to encompass all neurons in the brain in a variety of intersecting patterns. Overlapping 3-kb DNA fragments from the flanking noncoding and intronic regions of genes thought to have patterned expression in the adult brain were inserted into a defined genomic location by site-specific recombination. These fragments were then assayed for their ability to function as transcriptional enhancers in conjunction with a synthetic core promoter designed to work with a wide variety of enhancer types. An analysis of 44 fragments from four genes found that >80% drive expression patterns in the brain; the observed patterns were, on average, comprised of <100 cells. Our results suggest that the D. melanogaster genome contains >50,000 enhancers and that multiple enhancers drive distinct subsets of expression of a gene in each tissue and developmental stage. We expect that these lines will be valuable tools for neuroanatomy as well as for the elucidation of neuronal circuits and information flow in the fly brain.
In neurons, individual dendritic spines isolate N-methyl-d-aspartate (NMDA) receptor-mediated calcium ion (Ca2+) accumulations from the dendrite and other spines. However, the extent to which spines compartmentalize signaling events downstream of Ca2+ influx is not known. We combined two-photon fluorescence lifetime imaging with two-photon glutamate uncaging to image the activity of the small guanosine triphosphatase Ras after NMDA receptor activation at individual spines. Induction of long-term potentiation (LTP) triggered robust Ca2+-dependent Ras activation in single spines that decayed in approximately 5 minutes. Ras activity spread over approximately 10 micrometers of dendrite and invaded neighboring spines by diffusion. The spread of Ras-dependent signaling was necessary for the local regulation of the threshold for LTP induction. Thus, Ca2+-dependent synaptic signals can spread to couple multiple synapses on short stretches of dendrite.
Fluorescent proteins and their engineered variants have played an important role in the study of biology. The genetically encoded calcium-indicator protein GCaMP2 comprises a circularly permuted fluorescent protein coupled to the calcium-binding protein calmodulin and a calmodulin target peptide, M13, derived from the intracellular calmodulin target myosin light-chain kinase and has been used to image calcium transients in vivo. To aid rational efforts to engineer improved variants of GCaMP2, this protein was crystallized in the calcium-saturated form. X-ray diffraction data were collected to 2.0 A resolution. The crystals belong to space group C2, with unit-cell parameters a = 126.1.
We present a Bayesian framework for action recognition through ballistic dynamics. Psycho-kinesiological studies indicate that ballistic movements form the natural units for human movement planning. The framework leads to an efficient and robust algorithm for temporally segmenting videos into atomic movements. Individual movements are annotated with person-centric morphological labels called ballistic verbs. This is tested on a dataset of interactive movements, achieving high recognition rates. The approach is also applied on a gesture recognition task, improving a previously reported recognition rate from 84% to 92%. Consideration of ballistic dynamics enhances the performance of the popular Motion History Image feature. We also illustrate the approach’s general utility on real-world videos. Experiments indicate that the method is robust to view, style and appearance variations.
Sequence database searches require accurate estimation of the statistical significance of scores. Optimal local sequence alignment scores follow Gumbel distributions, but determining an important parameter of the distribution (lambda) requires time-consuming computational simulation. Moreover, optimal alignment scores are less powerful than probabilistic scores that integrate over alignment uncertainty ("Forward" scores), but the expected distribution of Forward scores remains unknown. Here, I conjecture that both expected score distributions have simple, predictable forms when full probabilistic modeling methods are used. For a probabilistic model of local sequence alignment, optimal alignment bit scores ("Viterbi" scores) are Gumbel-distributed with constant lambda = log 2, and the high scoring tail of Forward scores is exponential with the same constant lambda. Simulation studies support these conjectures over a wide range of profile/sequence comparisons, using 9,318 profile-hidden Markov models from the Pfam database. This enables efficient and accurate determination of expectation values (E-values) for both Viterbi and Forward scores for probabilistic local alignments.
Over hundreds of millions of years, evolution has optimized brain design to maximize its functionality while minimizing costs associated with building and maintenance. This observation suggests that one can use optimization theory to rationalize various features of brain design. Here, we attempt to explain the dimensions and branching structure of dendritic arbors by minimizing dendritic cost for given potential synaptic connectivity. Assuming only that dendritic cost increases with total dendritic length and path length from synapses to soma, we find that branching, planar, and compact dendritic arbors, such as those belonging to Purkinje cells in the cerebellum, are optimal. The theory predicts that adjacent Purkinje dendritic arbors should spatially segregate. In addition, we propose two explicit cost function expressions, falsifiable by measuring dendritic caliber near bifurcations.
We demonstrate live-cell super-resolution imaging using photoactivated localization microscopy (PALM). The use of photon-tolerant cell lines in combination with the high resolution and molecular sensitivity of PALM permitted us to investigate the nanoscale dynamics within individual adhesion complexes (ACs) in living cells under physiological conditions for as long as 25 min, with half of the time spent collecting the PALM images at spatial resolutions down to approximately 60 nm and frame rates as short as 25 s. We visualized the formation of ACs and measured the fractional gain and loss of individual paxillin molecules as each AC evolved. By allowing observation of a wide variety of nanoscale dynamics, live-cell PALM provides insights into molecular assembly during the initiation, maturation and dissolution of cellular processes.
Commentary: The first example of true live cell and time lapse imaging by localization microscopy (as opposed to particle tracking), this paper uses the Nyquist criterion to establish a necessary condition for true spatial resolution based on the density of localized molecules – a condition often unmet in claims elsewhere in the superresolution literature.
By any method, higher spatiotemporal resolution requires increasing light exposure at the specimen, making noninvasive imaging increasingly difficult. Here, simultaneous differential interference contrast imaging is used to establish that cells behave physiologically before, during, and after PALM imaging. Similar controls are lacking from many supposed “live cell” superresolution demonstrations.
The development of high-resolution microscopy makes possible the high-throughput screening of cellular information, such as gene expression at single cell resolution. One of the critical enabling techniques yet to be developed is the automatic recognition or annotation of specific cells in a 3D image stack. In this paper, we present a novel graph-based algorithm, ARC, that determines cell identities in a 3D confocal image of C. elegans based on their highly stereotyped arrangement. This is an essential step in our work on gene expression analysis of C. elegans at the resolution of single cells. Our ARC method integrates both the absolute and relative spatial locations of cells in a C. elegans body. It uses a marker-guided, spatially-constrained, two-stage bipartite matching to find the optimal match between cells in a subject image and cells in 15 template images that have been manually annotated and vetted. We applied ARC to the recognition of cells in 3D confocal images of the first larval stage (L1) of C. elegans hermaphrodites, and achieved an average accuracy of 94.91%.
Although information storage in the central nervous system is thought to be primarily mediated by various forms of synaptic plasticity, other mechanisms, such as modifications in membrane excitability, are available. Local dendritic spikes are nonlinear voltage events that are initiated within dendritic branches by spatially clustered and temporally synchronous synaptic input. That local spikes selectively respond only to appropriately correlated input allows them to function as input feature detectors and potentially as powerful information storage mechanisms. However, it is currently unknown whether any effective form of local dendritic spike plasticity exists. Here we show that the coupling between local dendritic spikes and the soma of rat hippocampal CA1 pyramidal neurons can be modified in a branch-specific manner through an N-methyl-d-aspartate receptor (NMDAR)-dependent regulation of dendritic Kv4.2 potassium channels. These data suggest that compartmentalized changes in branch excitability could store multiple complex features of synaptic input, such as their spatio-temporal correlation. We propose that this ’branch strength potentiation’ represents a previously unknown form of information storage that is distinct from that produced by changes in synaptic efficacy both at the mechanistic level and in the type of information stored.